Chromosome errors in human eggs shape natural fertility over reproductive life span
- PMID: 31604276
- PMCID: PMC7212007
- DOI: 10.1126/science.aav7321
Chromosome errors in human eggs shape natural fertility over reproductive life span
Abstract
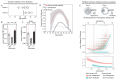
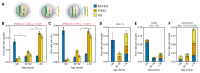
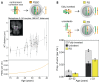
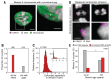
Chromosome errors, or aneuploidy, affect an exceptionally high number of human conceptions, causing pregnancy loss and congenital disorders. Here, we have followed chromosome segregation in human oocytes from females aged 9 to 43 years and report that aneuploidy follows a U-curve. Specific segregation error types show different age dependencies, providing a quantitative explanation for the U-curve. Whole-chromosome nondisjunction events are preferentially associated with increased aneuploidy in young girls, whereas centromeric and more extensive cohesion loss limit fertility as women age. Our findings suggest that chromosomal errors originating in oocytes determine the curve of natural fertility in humans.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures
Comment in
-
Re: Chromosome Errors in Human Eggs Shape Natural Fertility over Reproductive Life Span.J Urol. 2020 Jun;203(6):1066. doi: 10.1097/JU.0000000000001002.02. Epub 2020 Mar 10. J Urol. 2020. PMID: 32155374 No abstract available.
-
Advances of aneuploidy research in the maternal germline.Nat Rev Genet. 2023 May;24(5):274. doi: 10.1038/s41576-022-00573-7. Nat Rev Genet. 2023. PMID: 36635408 No abstract available.
References
-
- Kumar A, et al. Indian J Pediatr. 2007;74:927–931. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

