Comparative Genomics and Transcriptomics To Analyze Fruiting Body Development in Filamentous Ascomycetes
- PMID: 31604798
- PMCID: PMC6893386
- DOI: 10.1534/genetics.119.302749
Comparative Genomics and Transcriptomics To Analyze Fruiting Body Development in Filamentous Ascomycetes
Abstract
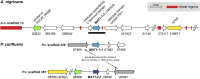
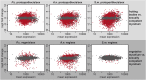
Many filamentous ascomycetes develop three-dimensional fruiting bodies for production and dispersal of sexual spores. Fruiting bodies are among the most complex structures differentiated by ascomycetes; however, the molecular mechanisms underlying this process are insufficiently understood. Previous comparative transcriptomics analyses of fruiting body development in different ascomycetes suggested that there might be a core set of genes that are transcriptionally regulated in a similar manner across species. Conserved patterns of gene expression can be indicative of functional relevance, and therefore such a set of genes might constitute promising candidates for functional analyses. In this study, we have sequenced the genome of the Pezizomycete Ascodesmis nigricans, and performed comparative transcriptomics of developing fruiting bodies of this fungus, the Pezizomycete Pyronema confluens, and the Sordariomycete Sordaria macrospora With only 27 Mb, the A. nigricans genome is the smallest Pezizomycete genome sequenced to date. Comparative transcriptomics indicated that gene expression patterns in developing fruiting bodies of the three species are more similar to each other than to nonsexual hyphae of the same species. An analysis of 83 genes that are upregulated only during fruiting body development in all three species revealed 23 genes encoding proteins with predicted roles in vesicle transport, the endomembrane system, or transport across membranes, and 13 genes encoding proteins with predicted roles in chromatin organization or the regulation of gene expression. Among four genes chosen for functional analysis by deletion in S. macrospora, three were shown to be involved in fruiting body formation, including two predicted chromatin modifier genes.
Keywords: Ascodesmis nigricans; Pyronema confluens; Sordaria macrospora; comparative transcriptomics; fruiting body development.
Copyright © 2019 by the Genetics Society of America.
Figures





References
-
- Al-Shahrour F., Minguez P., Tárraga J., Medina I., Alloza E. et al. , 2007. FatiGO +: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 35: W91–W96. 10.1093/nar/gkm260 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

