High-potency ligands for DREADD imaging and activation in rodents and monkeys
- PMID: 31604917
- PMCID: PMC6788984
- DOI: 10.1038/s41467-019-12236-z
High-potency ligands for DREADD imaging and activation in rodents and monkeys
Abstract
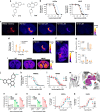
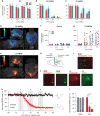
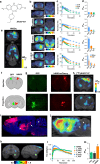
Designer Receptors Exclusively Activated by Designer Drugs (DREADDs) are a popular chemogenetic technology for manipulation of neuronal activity in uninstrumented awake animals with potential for human applications as well. The prototypical DREADD agonist clozapine N-oxide (CNO) lacks brain entry and converts to clozapine, making it difficult to apply in basic and translational applications. Here we report the development of two novel DREADD agonists, JHU37152 and JHU37160, and the first dedicated 18F positron emission tomography (PET) DREADD radiotracer, [18F]JHU37107. We show that JHU37152 and JHU37160 exhibit high in vivo DREADD potency. [18F]JHU37107 combined with PET allows for DREADD detection in locally-targeted neurons, and at their long-range projections, enabling noninvasive and longitudinal neuronal projection mapping.
Conflict of interest statement
M.M. is a cofounder and owns stock in Metis Laboratories. J.B., J.L.G., F.H., M.S.S., A.G.H., M.G.P., and M.M. are listed as inventors on an application (62/627,527) filed with the U.S. Patent Office regarding the novel DREADD compounds described herein. Remaining authors declare no competing interest.
Figures

References
-
- Thompson Karen J., Khajehali Elham, Bradley Sophie J., Navarrete Jovana S., Huang Xi Ping, Slocum Samuel, Jin Jian, Liu Jing, Xiong Yan, Olsen Reid H. J., Diberto Jeffrey F., Boyt Kristen M., Pina Melanie M., Pati Dipanwita, Molloy Colin, Bundgaard Christoffer, Sexton Patrick M., Kash Thomas L., Krashes Michael J., Christopoulos Arthur, Roth Bryan L., Tobin Andrew B. DREADD Agonist 21 Is an Effective Agonist for Muscarinic-Based DREADDs in Vitro and in Vivo. ACS Pharmacology & Translational Science. 2018;1(1):61–72. doi: 10.1021/acsptsci.8b00012. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ZIA MH002793/MH/NIMH NIH HHS/United States
- ZIA MH002795/ImNIH/Intramural NIH HHS/United States
- ZIA000069/U.S. Department of Health & Human Services | NIH | National Institute on Drug Abuse (NIDA)/International
- ZIA MH002795/MH/NIMH NIH HHS/United States
- ZIA MH002793/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

