Contribution of retrotransposition to developmental disorders
- PMID: 31604926
- PMCID: PMC6789007
- DOI: 10.1038/s41467-019-12520-y
Contribution of retrotransposition to developmental disorders
Abstract
Mobile genetic Elements (MEs) are segments of DNA which can copy themselves and other transcribed sequences through the process of retrotransposition (RT). In humans several disorders have been attributed to RT, but the role of RT in severe developmental disorders (DD) has not yet been explored. Here we identify RT-derived events in 9738 exome sequenced trios with DD-affected probands. We ascertain 9 de novo MEs, 4 of which are likely causative of the patient's symptoms (0.04%), as well as 2 de novo gene retroduplications. Beyond identifying likely diagnostic RT events, we estimate genome-wide germline ME mutation rate and selective constraint and demonstrate that coding RT events have signatures of purifying selection equivalent to those of truncating mutations. Overall, our analysis represents a comprehensive interrogation of the impact of retrotransposition on protein coding genes and a framework for future evolutionary and disease studies.
Conflict of interest statement
M.E.H. is a co-founder of, consultant to, and holds shares in, Congenica Ltd, a genetics diagnostic company. The remaining authors declare no competing interests.
Figures
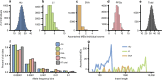

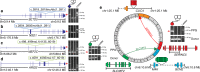

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

