Homocysteine-methionine cycle is a metabolic sensor system controlling methylation-regulated pathological signaling
- PMID: 31605963
- PMCID: PMC6812029
- DOI: 10.1016/j.redox.2019.101322
Homocysteine-methionine cycle is a metabolic sensor system controlling methylation-regulated pathological signaling
Abstract
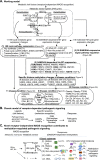
Homocysteine-Methionine (HM) cycle produces universal methyl group donor S-adenosylmethione (SAM), methyltransferase inhibitor S-adenosylhomocysteine (SAH) and homocysteine (Hcy). Hyperhomocysteinemia (HHcy) is established as an independent risk factor for cardiovascular disease (CVD) and other degenerative disease. We selected 115 genes in the extended HM cycle (31 metabolic enzymes and 84 methyltransferases), examined their protein subcellular location/partner protein, investigated their mRNA levels and mapped their corresponding histone methylation status in 35 disease conditions via mining a set of public databases and intensive literature research. We have 6 major findings. 1) All HM metabolic enzymes are located only in the cytosol except for cystathionine-β-synthase (CBS), which was identified in both cytosol and nucleus. 2) Eight disease conditions encountered only histone hypomethylation on 8 histone residues (H3R2/K4/R8/K9/K27/K36/K79 and H4R3). Nine disease conditions had only histone hypermethylation on 8 histone residues (H3R2/K4/K9/K27/K36/K79 and H4R3/K20). 3) We classified 9 disease types with differential HM cycle expression pattern. Eleven disease conditions presented most 4 HM cycle pathway suppression. 4) Three disease conditions had all 4 HM cycle pathway suppression and only histone hypomethylation on H3R2/K4/R8/K9/K36 and H4R3. 5) Eleven HM cycle metabolic enzymes interact with 955 proteins. 6) Five paired HM cycle proteins interact with each other. We conclude that HM cycle is a key metabolic sensor system which mediates receptor-independent metabolism-associated danger signal recognition and modulates SAM/SAH-dependent methylation in disease conditions and that hypomethylation on frequently modified histone residues is a key mechanism for metabolic disorders, autoimmune disease and CVD. We propose that HM metabolism takes place in the cytosol, that nuclear methylation equilibration requires a nuclear-cytosol transfer of SAM/SAH/Hcy, and that Hcy clearance is essential for genetic protection.
Keywords: Homocysteine-methionine cycle; Metabolic sensor; SAM/SAH-dependent methylation.
Copyright © 2019. Published by Elsevier B.V.
Figures





Similar articles
-
Regulation of homocysteine metabolism and methylation in human and mouse tissues.FASEB J. 2010 Aug;24(8):2804-17. doi: 10.1096/fj.09-143651. Epub 2010 Mar 19. FASEB J. 2010. PMID: 20305127 Free PMC article.
-
Hyperhomocysteinemia and response of methionine cycle intermediates to vitamin treatment in renal patients.Clin Chem Lab Med. 2005;43(10):1039-47. doi: 10.1515/CCLM.2005.182. Clin Chem Lab Med. 2005. PMID: 16197295 Review.
-
The nutrigenetics of hyperhomocysteinemia: quantitative proteomics reveals differences in the methionine cycle enzymes of gene-induced versus diet-induced hyperhomocysteinemia.Mol Cell Proteomics. 2010 Mar;9(3):471-85. doi: 10.1074/mcp.M900406-MCP200. Epub 2009 Dec 14. Mol Cell Proteomics. 2010. PMID: 20008833 Free PMC article.
-
The Contribution of Homocysteine Metabolism Disruption to Endothelial Dysfunction: State-of-the-Art.Int J Mol Sci. 2019 Feb 17;20(4):867. doi: 10.3390/ijms20040867. Int J Mol Sci. 2019. PMID: 30781581 Free PMC article. Review.
-
Global protein and histone arginine methylation are affected in a tissue-specific manner in a rat model of diet-induced hyperhomocysteinemia.Biochim Biophys Acta. 2013 Oct;1832(10):1708-14. doi: 10.1016/j.bbadis.2013.05.013. Epub 2013 May 22. Biochim Biophys Acta. 2013. PMID: 23707560
Cited by
-
Innate immunity of vascular smooth muscle cells contributes to two-wave inflammation in atherosclerosis, twin-peak inflammation in aortic aneurysms and trans-differentiation potential into 25 cell types.Front Immunol. 2024 Jan 24;14:1348238. doi: 10.3389/fimmu.2023.1348238. eCollection 2023. Front Immunol. 2024. PMID: 38327764 Free PMC article.
-
Relationship of Homocysteine Plasma Levels with Mild Cognitive Impairment, Alzheimer's Disease, Vascular Dementia, Psychobehavioral, and Functional Complications.J Alzheimers Dis. 2021;82(1):235-248. doi: 10.3233/JAD-210166. J Alzheimers Dis. 2021. PMID: 34057086 Free PMC article.
-
Effect of a Phytochemical-Rich Olive-Derived Extract on Anthropometric, Hematological, and Metabolic Parameters.Nutrients. 2024 Sep 11;16(18):3068. doi: 10.3390/nu16183068. Nutrients. 2024. PMID: 39339668 Free PMC article.
-
Serum folate concentration and health-related quality of life among the elderly in South Korea.Health Qual Life Outcomes. 2021 Dec 20;19(1):267. doi: 10.1186/s12955-021-01899-2. Health Qual Life Outcomes. 2021. PMID: 34930296 Free PMC article.
-
Examination of Genetic and Epigenetic Characteristics of Patients with Hyperhomocysteinemia Following High-Dose Folic Acid Consumption.Nutrients. 2025 Jun 27;17(13):2133. doi: 10.3390/nu17132133. Nutrients. 2025. PMID: 40647238 Free PMC article.
References
-
- Jensen M.K. Novel metabolic biomarkers of cardiovascular disease. Nat. Rev. Endocrinol. 2014;10(11):659–672. - PubMed
-
- Martinez E., Martorell J., Riambau V. Review of serum biomarkers in carotid atherosclerosis. J. Vasc. Surg. 2019 - PubMed
-
- Feinberg A.P., Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4(2):143–153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

