Analysis of cardiac troponin proteoforms by top-down mass spectrometry
- PMID: 31606082
- PMCID: PMC7557121
- DOI: 10.1016/bs.mie.2019.07.029
Analysis of cardiac troponin proteoforms by top-down mass spectrometry
Abstract
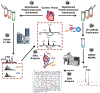
The cardiac troponin complex, composed of three regulatory proteins (cTnI, cTnT, TnC), functions as the critical regulator of cardiac muscle contraction and relaxation. Myofilament protein-protein interactions are regulated by post-translational modifications (PTMs) to the protein constituents of this complex. Dysregulation of troponin PTMs, particularly phosphorylation, results in altered cardiac contractility. Altered PTMs and isoforms have been increasingly recognized as the molecular mechanisms underlying heart diseases. Therefore, it is essential to comprehensively analyze cardiac troponin proteoforms that arise from PTMs, alternative splicing, and sequence variations. In this chapter, we described two detailed protocols for the enrichment and purification of endogenous cardiac troponin proteoforms from cardiac tissue. Subsequently, mass spectrometry (MS)-based top-down proteomics utilizing online liquid chromatography (LC)/quadrupole time-of-flight (Q-TOF) MS for separation, profiling, and quantification of the troponins was demonstrated. Characterization of troponin amino acid sequence and the localization of PTMs were shown using Fourier-transform ion cyclotron resonance (FT-ICR) MS with electron capture dissociation (ECD) and collisionally activated dissociation (CAD). Furthermore, we described the use of MASH software, a comprehensive and free software package developed in our lab, for top-down proteomics data analysis. The methods we described can be applied for the analysis of troponin proteoforms in cardiac tissues, from animal models to human clinical samples, for heart disease.
Keywords: Cardiac troponin proteoforms; Phosphorylation; Protein extraction; Top-down proteomics.
© 2019 Elsevier Inc. All rights reserved.
Figures



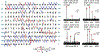

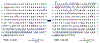
References
-
- Ansong C, Wu S, Meng D, Liu X, Brewer HM, Deatherage Kaiser BL, … Pasa-Tolic L. (2013). Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proceedings of the National Academy of Sciences, 110, 10153–10158. - PMC - PubMed
-
- Apple FS, Sandoval Y, Jaffe AS, & Ordonez-Llanos J. (2017). Cardiac Troponin Assays: Guide to Understanding Analytical Characteristics and Their Impact on Clinical Care. Clinical Chemistry, 63, 73–81. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

