Conserved aging-related signatures of senescence and inflammation in different tissues and species
- PMID: 31606727
- PMCID: PMC6814591
- DOI: 10.18632/aging.102345
Conserved aging-related signatures of senescence and inflammation in different tissues and species
Abstract
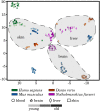
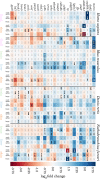
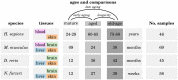
Increasing evidence indicates that chronic inflammation and senescence are the cause of many severe age-related diseases, with both biological processes highly upregulated during aging. However, until now, it has remained unknown whether specific inflammation- or senescence-related genes exist that are common between different species or tissues. These potential markers of aging could help to identify possible targets for therapeutic interventions of aging-associated afflictions and might also deepen our understanding of the principal mechanisms of aging. With the objective of identifying such signatures of aging and tissue-specific aging markers, we analyzed a multitude of cross-sectional RNA-Seq data from four evolutionarily distinct species (human, mouse and two fish) and four different tissues (blood, brain, liver and skin). In at least three different species and three different tissues, we identified several genes that displayed similar expression patterns that might serve as potential aging markers. Additionally, we show that genes involved in aging-related processes tend to be tighter controlled in long-lived than in average-lived individuals. These observations hint at a general genetic level that affect an individual's life span. Altogether, this descriptive study contributes to a better understanding of common aging signatures as well as tissue-specific aging patterns and supplies the basis for further investigative age-related studies.
Keywords: RNA-Seq; aging; inflammaging; senescence; transcriptomics.
Conflict of interest statement
Figures



References
-
- Aramillo Irizar P, Schäuble S, Esser D, Groth M, Frahm C, Priebe S, Baumgart M, Hartmann N, Marthandan S, Menzel U, Müller J, Schmidt S, Ast V, et al.. Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly. Nat Commun. 2018; 9:327. 10.1038/s41467-017-02395-2 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

