Human Cortical Organoids Expose a Differential Function of GSK3 on Cortical Neurogenesis
- PMID: 31607568
- PMCID: PMC6893153
- DOI: 10.1016/j.stemcr.2019.09.005
Human Cortical Organoids Expose a Differential Function of GSK3 on Cortical Neurogenesis
Abstract
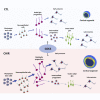
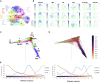
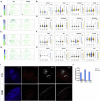
The regulation of the proliferation and polarity of neural progenitors is crucial for the development of the brain cortex. Animal studies have implicated glycogen synthase kinase 3 (GSK3) as a pivotal regulator of both proliferation and polarity, yet the functional relevance of its signaling for the unique features of human corticogenesis remains to be elucidated. We harnessed human cortical brain organoids to probe the longitudinal impact of GSK3 inhibition through multiple developmental stages. Chronic GSK3 inhibition increased the proliferation of neural progenitors and caused massive derangement of cortical tissue architecture. Single-cell transcriptome profiling revealed a direct impact on early neurogenesis and uncovered a selective role of GSK3 in the regulation of glutamatergic lineages and outer radial glia output. Our dissection of the GSK3-dependent transcriptional network in human corticogenesis underscores the robustness of the programs determining neuronal identity independent of tissue architecture.
Keywords: GSK3; corticogenesis; human brain organoids; outer radial glia; single cell transcriptomics.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Beattie R., Postiglione M.P., Burnett L.E., Laukoter S., Streicher C., Pauler F.M., Xiao G., Klezovitch O., Vasioukhin V., Ghashghaei T.H. Mosaic analysis with double markers reveals distinct sequential functions of Lgl1 in neural stem cells. Neuron. 2017;94:517–533.e3. - PubMed
-
- Betizeau M., Cortay V., Patti D., Pfister S., Gautier E., Bellemin-Ménard A., Afanassieff M., Huissoud C., Douglas R.J., Kennedy H. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

