Diel Oscillation of Microbial Gene Transcripts Declines With Depth in Oligotrophic Ocean Waters
- PMID: 31608031
- PMCID: PMC6769238
- DOI: 10.3389/fmicb.2019.02191
Diel Oscillation of Microbial Gene Transcripts Declines With Depth in Oligotrophic Ocean Waters
Abstract
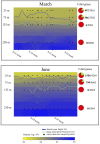
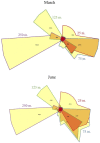
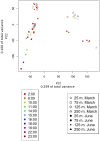
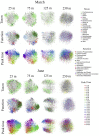
Diel oscillations in primary and secondary production, growth, metabolic activity, and gene expression commonly occur in marine microbial communities in ocean surface waters. Diel periodicity of gene transcription has been demonstrated in photoautotrophic and heterotrophic microbes in both coastal and open ocean environments. To better define the spatiotemporal distribution and patterns of these daily oscillations, we investigated how diel periodicity in gene transcripts changed with depth from the surface waters to the upper mesopelagic. We postulated that diel oscillation of transcript abundances would diminish at greater depths across the collective microbial community due to decreasing light availability. The results showed that the number and total proportion of gene transcripts and taxa exhibiting diel periodicity were greatest in the shallow sunlit mixed layer, diminished rapidly with increasing depth to the base of the euphotic zone, and could not be detected in the mesopelagic. The results confirmed an overall decrease in microbial diel transcript oscillation with depth through the euphotic zone and suggested a relationship between abundance of diel oscillating transcripts and the daily integrated light exposure experienced by planktonic microbes in the water column. Local dissolved macronutrient concentration also appeared to influence the diel transcriptional patterns of specific microbial genes. The diminishing diel transcript oscillations found at increasing depths suggest that diel patterns of other microbial processes and interactions may likewise be attenuated at depth.
Keywords: bacterioplankton; diel; oceanography; oligotrophic; phytoplankton; transcriptome.
Copyright © 2019 Vislova, Sosa, Eppley, Romano and DeLong.
Figures

References
Associated data
LinkOut - more resources
Full Text Sources

