Hsa_circ_0000479 as a Novel Diagnostic Biomarker of Systemic Lupus Erythematosus
- PMID: 31608065
- PMCID: PMC6771011
- DOI: 10.3389/fimmu.2019.02281
Hsa_circ_0000479 as a Novel Diagnostic Biomarker of Systemic Lupus Erythematosus
Abstract
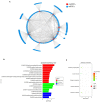
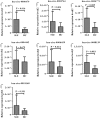
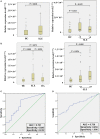
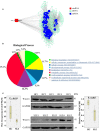
Background: Accumulating evidence suggests that differentially expressed non-coding circular RNAs (circRNAs) play critical roles in the progress of autoimmune diseases. However, the role of circRNAs in systemic lupus erythematosus (SLE) remains unclear. Methods: We initially used next-generation sequencing (NGS) to comprehensively analyze circRNA expression profiles in peripheral blood mononuclear cells (PBMCs) from 10 SLE patients, stratified by their disease activity characteristics (stable or active SLE), and 10 healthy controls (HCs). Candidate circRNAs identified were first validated by quantitative reverse-transcription (qRT)-PCR in PBMC samples from a training-phase cohort of five SLE patients and five HCs. The significantly dysregulated circRNAs were then confirmed by qRT-PCR in a validation cohort of 23 SLE patients and 21 HCs, and in an external validation cohort with 64 SLE patients, 58 HCs, and 50 patients with rheumatoid arthritis (RA). In addition, we conducted bioinformatics analysis and western blotting investigating the relationships between the candidate circRNAs and SLE progression. Results: Multilayer integrative analysis of circRNA regulation showed that 84 circRNAs were upregulated and 30 were downregulated in patients with SLE compared with HCs. We then analyzed the intersection of these differentially expressed circRNAs in an SLE-stable cohort, an SLE-active cohort, and HCs. This enabled us to narrow down dysregulated circRNAs to 15 upregulated circRNAs. Only hsa_circ_0000479 was significantly upregulated in PBMCs of patients with SLE compared with HCs (P < 0.05). Furthermore, the diagnostic potential of hsa_circ_0000479 expression to distinguish SLE patients from HCs and RA patients was also significantly increased in the validation-phase and external-validation-phase cohorts (P < 0.05). When distinguishing SLE patients from HCs, the diagnostic specificities of hsa_circ_0000479 were 0.619 and 1.0 in two validation cohorts, respectively (AUCs = 0.731 and 0.730, respectively). It was also significantly increased in either stable SLE patients or active SLE patients compared with HCs in these two cohorts (P < 0.05). We also used bioinformatics analysis to show that hsa_circ_0000479 regulates SLE progression by modulating metabolic pathways and the Wnt signaling pathway. Western blotting revealed that the expression of Wnt-16 protein was significantly decreased in SLE. Conclusion: Our results suggest that hsa_circ_0000479 has potential as a novel biomarker for the diagnosis of SLE.
Keywords: RNA-sequencing; biomarker; circular RNAs (circRNAs); hsa_circ_0000479; systemic lupus erythematosus.
Copyright © 2019 Guo, Wang, Ye, Shi, Yan, Lin, Huang, Li, Lin, Zhu, Xue and Zhang.
Figures

References
-
- Zhang H, Huang X, Ye L, Guo G, Li X, Chen C, et al. . B cell-related circulating MicroRNAs with the potential value of biomarkers in the differential diagnosis, and distinguishment between the disease activity and lupus nephritis for systemic lupus erythematosus. Front Immunol. (2018) 9:1473. 10.3389/fimmu.2018.01473 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

