Elimination of Reference Mapping Bias Reveals Robust Immune Related Allele-Specific Expression in Crossbred Sheep
- PMID: 31608110
- PMCID: PMC6761296
- DOI: 10.3389/fgene.2019.00863
Elimination of Reference Mapping Bias Reveals Robust Immune Related Allele-Specific Expression in Crossbred Sheep
Abstract
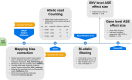
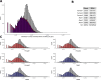
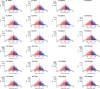
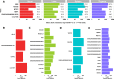
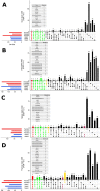
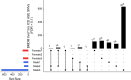
Pervasive allelic variation at both gene and single nucleotide level (SNV) between individuals is commonly associated with complex traits in humans and animals. Allele-specific expression (ASE) analysis, using RNA-Seq, can provide a detailed annotation of allelic imbalance and infer the existence of cis-acting transcriptional regulation. However, variant detection in RNA-Seq data is compromised by biased mapping of reads to the reference DNA sequence. In this manuscript, we describe an unbiased standardized computational pipeline for allele-specific expression analysis using RNA-Seq data, which we have adapted and developed using tools available under open license. The analysis pipeline we present is designed to minimize reference bias while providing accurate profiling of allele-specific expression across tissues and cell types. Using this methodology, we were able to profile pervasive allelic imbalance across tissues and cell types, at both the gene and SNV level, in Texel×Scottish Blackface sheep, using the sheep gene expression atlas data set. ASE profiles were pervasive in each sheep and across all tissue types investigated. However, ASE profiles shared across tissues were limited, and instead, they tended to be highly tissue-specific. These tissue-specific ASE profiles may underlie the expression of economically important traits and could be utilized as weighted SNVs, for example, to improve the accuracy of genomic selection in breeding programs for sheep. An additional benefit of the pipeline is that it does not require parental genotypes and can therefore be applied to other RNA-Seq data sets for livestock, including those available on the Functional Annotation of Animal Genomes (FAANG) data portal. This study is the first global characterization of moderate to extreme ASE in tissues and cell types from sheep. We have applied a robust methodology for ASE profiling to provide both a novel analysis of the multi-dimensional sheep gene expression atlas data set and a foundation for identifying the regulatory and expressed elements of the genome that are driving complex traits in livestock.
Keywords: GeneiASE; RNA-Seq; WASP; allele-specific expression; mapping bias; sheep; transcriptome.
Copyright © 2019 Salavati, Bush, Palma-Vera, McCulloch, Hume and Clark.
Figures

References
-
- Álvarez I., Pérez-Pardal L., Traoré A., Fernández I., Goyache F. (2016). Lack of specific alleles for the bovine chemokine (C-X-C) receptor type 4 (CXCR4) gene in West African cattle questions its role as a candidate for trypanotolerance. Infect. Genet. Evol. 42, 30–33. 10.1016/j.meegid.2016.04.029 - DOI - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing on JSTOR. Source J. R. Stat. Soc. Ser. B 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

