Advances and Challenges in Metatranscriptomic Analysis
- PMID: 31608125
- PMCID: PMC6774269
- DOI: 10.3389/fgene.2019.00904
Advances and Challenges in Metatranscriptomic Analysis
Abstract
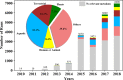
Sequencing-based analyses of microbiomes have traditionally focused on addressing the question of community membership and profiling taxonomic abundance through amplicon sequencing of 16 rRNA genes. More recently, shotgun metagenomics, which involves the random sequencing of all genomic content of a microbiome, has dominated this arena due to advancements in sequencing technology throughput and capability to profile genes as well as microbiome membership. While these methods have revealed a great number of insights into a wide variety of microbiomes, both of these approaches only describe the presence of organisms or genes, and not whether they are active members of the microbiome. To obtain deeper insights into how a microbial community responds over time to their changing environmental conditions, microbiome scientists are beginning to employ large-scale metatranscriptomics approaches. Here, we present a comprehensive review on computational metatranscriptomics approaches to study microbial community transcriptomes. We review the major advancements in this burgeoning field, compare strengths and weaknesses to other microbiome analysis methods, list available tools and workflows, and describe use cases and limitations of this method. We envision that this field will continue to grow exponentially, as will the scope of projects (e.g. longitudinal studies of community transcriptional responses to perturbations over time) and the resulting data. This review will provide a list of options for computational analysis of these data and will highlight areas in need of development.
Keywords: RNASeq; gene expression; microbiome; omics; workflows.
Copyright © 2019 Shakya, Lo and Chain.
Figures
References
-
- Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data.
-
- Berg C., Dupont C. L., Asplund-Samuelsson J., Celepli N. A., Eiler A., Allen A. E., et al. (2018). Dissection of microbial community functions during a cyanobacterial bloom in the baltic sea via metatranscriptomics. Front. Mar. Sci. 5, 55. 10.3389/fmars.2018.00055 - DOI
Publication types
LinkOut - more resources
Full Text Sources

