Mapping of a Regulatory Site of the Escherichia coli ADP-Glucose Pyrophosphorylase
- PMID: 31608288
- PMCID: PMC6773804
- DOI: 10.3389/fmolb.2019.00089
Mapping of a Regulatory Site of the Escherichia coli ADP-Glucose Pyrophosphorylase
Abstract
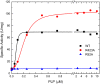
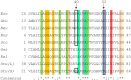
The enzyme ADP-glucose pyrophosphorylase (ADP-Glc PPase) controls the biosynthesis of glycogen in bacteria and starch in plants. It is regulated by various activators in different organisms according to their metabolic characteristics. In Escherichia coli, the major allosteric activator is fructose 1,6-bisphosphate (FBP). Other potent activator analogs include 1,6-hexanediol bisphosphate (HBP) and pyridoxal 5'-phosphate (PLP). Recently, a crystal structure with FBP bound was reported (PDB ID: 5L6S). However, it is possible that the FBP site found is not directly responsible for the activation of the enzyme. We hypothesized FBP activates by binding one of its phosphate groups to another site ("P1") in which a sulfate molecule was observed. In the E. coli enzyme, Arg40, Arg52, and Arg386 are part of this "P1" pocket and tightly complex this sulfate, which is also present in the crystal structures of ADP-Glc PPases from Agrobacterium tumefaciens and Solanum tuberosum. To test this hypothesis, we modeled alternative binding conformations of FBP, HBP, and PLP into "P1." In addition, we performed a scanning mutagenesis of Arg residues near potential phosphate binding sites ("P1," "P2," "P3"). We found that Arg40 and Arg52 are essential for FBP and PLP binding and activation. In addition, mutation of Arg386 to Ala decreased the apparent affinity for the activators more than 35-fold. We propose that the activator binds at this "P1" pocket, as well as "P2." Arg40 and Arg52 are highly conserved residues and they may be a common feature to complex the phosphate moiety of different sugar phosphate activators in the ADP-Glc PPase family.
Keywords: allosteric regulation; arginine scanning mutagenesis; polysaccharide biosynthesis; pyridoxal 5′-phosphate activation; sugar phosphate regulation.
Copyright © 2019 Bhayani, Hill, Sharma, Iglesias, Olsen and Ballicora.
Figures



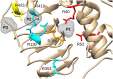
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

