Centromeric SMC1 promotes centromere clustering and stabilizes meiotic homolog pairing
- PMID: 31609962
- PMCID: PMC6812850
- DOI: 10.1371/journal.pgen.1008412
Centromeric SMC1 promotes centromere clustering and stabilizes meiotic homolog pairing
Abstract
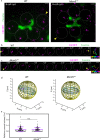
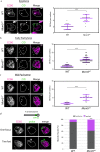
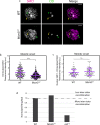
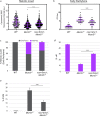
During meiosis, each chromosome must selectively pair and synapse with its own unique homolog to enable crossover formation and subsequent segregation. How homolog pairing is maintained in early meiosis to ensure synapsis occurs exclusively between homologs is unknown. We aimed to further understand this process by examining the meiotic defects of a unique Drosophila mutant, Mcm5A7. We found that Mcm5A7 mutants are proficient in homolog pairing at meiotic onset yet fail to maintain pairing as meiotic synapsis ensues, causing seemingly normal synapsis between non-homologous loci. This pairing defect corresponds with a reduction of SMC1-dependent centromere clustering at meiotic onset. Overexpressing SMC1 in this mutant significantly restores centromere clustering, homolog pairing, and crossover formation. These data indicate that the initial meiotic pairing of homologs is not sufficient to yield synapsis exclusively between homologs and provide a model in which meiotic homolog pairing must be stabilized by centromeric SMC1 to ensure proper synapsis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
The cohesion protein SOLO associates with SMC1 and is required for synapsis, recombination, homolog bias and cohesion and pairing of centromeres in Drosophila Meiosis.PLoS Genet. 2013;9(7):e1003637. doi: 10.1371/journal.pgen.1003637. Epub 2013 Jul 18. PLoS Genet. 2013. PMID: 23874232 Free PMC article.
-
Centromere pairing precedes meiotic chromosome pairing in plants.Sci China Life Sci. 2017 Nov;60(11):1197-1202. doi: 10.1007/s11427-017-9109-y. Epub 2017 Jul 26. Sci China Life Sci. 2017. PMID: 28755295 Review.
-
Oligopaint DNA FISH reveals telomere-based meiotic pairing dynamics in the silkworm, Bombyx mori.PLoS Genet. 2021 Jul 28;17(7):e1009700. doi: 10.1371/journal.pgen.1009700. eCollection 2021 Jul. PLoS Genet. 2021. PMID: 34319984 Free PMC article.
-
Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes.Curr Biol. 2011 Nov 8;21(21):1845-51. doi: 10.1016/j.cub.2011.09.044. Epub 2011 Oct 27. Curr Biol. 2011. PMID: 22036182
-
Female Meiosis: Synapsis, Recombination, and Segregation in Drosophila melanogaster.Genetics. 2018 Mar;208(3):875-908. doi: 10.1534/genetics.117.300081. Genetics. 2018. PMID: 29487146 Free PMC article. Review.
Cited by
-
A pathway for error-free non-homologous end joining of resected meiotic double-strand breaks.Nucleic Acids Res. 2021 Jan 25;49(2):879-890. doi: 10.1093/nar/gkaa1205. Nucleic Acids Res. 2021. PMID: 33406239 Free PMC article.
-
The RNA binding protein DND1 is elevated in a subpopulation of pro-spermatogonia and targets chromatin modifiers and translational machinery during late gestation.PLoS Genet. 2023 Mar 1;19(3):e1010656. doi: 10.1371/journal.pgen.1010656. eCollection 2023 Mar. PLoS Genet. 2023. PMID: 36857387 Free PMC article.
-
Centromeres: From chromosome biology to biotechnology applications and synthetic genomes in plants.Plant Biotechnol J. 2022 Nov;20(11):2051-2063. doi: 10.1111/pbi.13875. Epub 2022 Jul 7. Plant Biotechnol J. 2022. PMID: 35722725 Free PMC article. Review.
-
The Cohesin Complex Subunit ZmSMC3 Participates in Meiotic Centromere Pairing in Maize.Plant Cell. 2020 Apr;32(4):1323-1336. doi: 10.1105/tpc.19.00834. Epub 2020 Jan 29. Plant Cell. 2020. PMID: 31996400 Free PMC article.
-
PCH-2 collaborates with CMT-1 to proofread meiotic homolog interactions.PLoS Genet. 2020 Jul 30;16(7):e1008904. doi: 10.1371/journal.pgen.1008904. eCollection 2020 Jul. PLoS Genet. 2020. PMID: 32730253 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

