Molecular insights into the interaction of hemorphin and its targets
- PMID: 31611567
- PMCID: PMC6791854
- DOI: 10.1038/s41598-019-50619-w
Molecular insights into the interaction of hemorphin and its targets
Abstract
Hemorphins are atypical endogenous opioid peptides produced by the cleavage of hemoglobin beta chain. Several studies have reported the therapeutic potential of hemorphin in memory enhancement, blood regulation, and analgesia. However, the mode of interaction of hemorphin with its target remains largely elusive. The decapeptide LVV-hemorphin-7 is the most stable form of hemorphin. It binds with high affinity to mu-opioid receptors (MOR), angiotensin-converting enzyme (ACE) and insulin-regulated aminopeptidase (IRAP). In this study, computational methods were used extensively to elucidate the most likely binding pose of mammalian LVV-hemorphin-7 with the aforementioned proteins and to calculate the binding affinity. Additionally, alignment of mammalian hemorphin sequences showed that the hemorphin sequence of the camel harbors a variation - a Q > R substitution at position 8. This study also investigated the binding affinity and the interaction mechanism of camel LVV-hemorphin-7 with these proteins. To gain a better understanding of the dynamics of the molecular interactions between the selected targets and hemorphin peptides, 100 ns molecular dynamics simulations of the best-ranked poses were performed. Simulations highlighted major interactions between the peptides and key residues in the binding site of the proteins. Interestingly, camel hemorphin had a higher binding affinity and showed more interactions with all three proteins when compared to the canonical mammalian LVV-hemorphin-7. Thus, camel LVV-hemorphin-7 could be explored as a potent therapeutic agent for memory loss, hypertension, and analgesia.
Conflict of interest statement
The authors declare no competing interests.
Figures




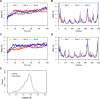
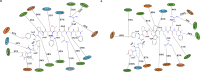


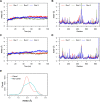

References
-
- Fruitier, I., Garreau, I. & Piot, J.-M. Cathepsin D Is a Good Candidate for the Specific Release of a Stable Hemorphin from Hemoglobin In Vivo: VV-Hemorphin-7. Biochem. Biophys. Res. Commun. 246, 719–724 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

