A new species in the major malaria vector complex sheds light on reticulated species evolution
- PMID: 31611571
- PMCID: PMC6791875
- DOI: 10.1038/s41598-019-49065-5
A new species in the major malaria vector complex sheds light on reticulated species evolution
Abstract
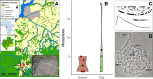
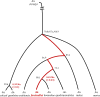
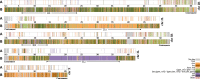
Complexes of closely related species provide key insights into the rapid and independent evolution of adaptive traits. Here, we described and studied Anopheles fontenillei sp.n., a new species in the Anopheles gambiae complex that we recently discovered in the forested areas of Gabon, Central Africa. Our analysis placed the new taxon in the phylogenetic tree of the An. gambiae complex, revealing important introgression events with other members of the complex. Particularly, we detected recent introgression, with Anopheles gambiae and Anopheles coluzzii, of genes directly involved in vectorial capacity. Moreover, genome analysis of the new species allowed us to clarify the evolutionary history of the 3La inversion. Overall, An. fontenillei sp.n. analysis improved our understanding of the relationship between species within the An. gambiae complex, and provided insight into the evolution of vectorial capacity traits that are relevant for the successful control of malaria in Africa.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Coyne, J. & Orr, H. Speciation. (Sinauer Associates, 2004).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

