Integrated nuclear proteomics and transcriptomics identifies S100A4 as a therapeutic target in acute myeloid leukemia
- PMID: 31611628
- PMCID: PMC6995695
- DOI: 10.1038/s41375-019-0596-4
Integrated nuclear proteomics and transcriptomics identifies S100A4 as a therapeutic target in acute myeloid leukemia
Abstract
Inappropriate localization of proteins can interfere with normal cellular function and drive tumor development. To understand how this contributes to the development of acute myeloid leukemia (AML), we compared the nuclear proteome and transcriptome of AML blasts with normal human CD34+ cells. Analysis of the proteome identified networks and processes that significantly affected transcription regulation including misexpression of 11 transcription factors with seven proteins not previously implicated in AML. Transcriptome analysis identified changes in 40 transcription factors but none of these were predictive of changes at the protein level. The highest differentially expressed protein in AML nuclei compared with normal CD34+ nuclei (not previously implicated in AML) was S100A4. In an extended cohort, we found that over-expression of nuclear S100A4 was highly prevalent in AML (83%; 20/24 AML patients). Knock down of S100A4 in AML cell lines strongly impacted their survival whilst normal hemopoietic stem progenitor cells were unaffected. These data are the first analysis of the nuclear proteome in AML and have identified changes in transcription factor expression or regulation of transcription that would not have been seen at the mRNA level. These data also suggest that S100A4 is essential for AML survival and could be a therapeutic target in AML.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
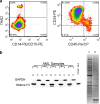


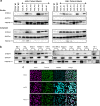

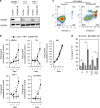
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

