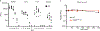Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence
- PMID: 31611642
- PMCID: PMC6988571
- DOI: 10.1038/s41564-019-0583-6
Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence
Abstract
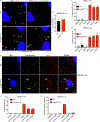
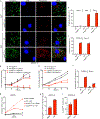
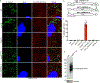
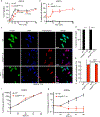
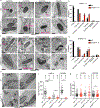
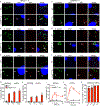
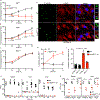
Rickettsia are obligate intracellular bacteria that evade antimicrobial autophagy in the host cell cytosol by unknown mechanisms. Other cytosolic pathogens block different steps of autophagy targeting, including the initial step of polyubiquitin-coat formation. One mechanism of evasion is to mobilize actin to the bacterial surface. Here, we show that actin mobilization is insufficient to block autophagy recognition of the pathogen Rickettsia parkeri. Instead, R. parkeri employs outer membrane protein B (OmpB) to block ubiquitylation of the bacterial surface proteins, including OmpA, and subsequent recognition by autophagy receptors. OmpB is also required for the formation of a capsule-like layer. Although OmpB is dispensable for bacterial growth in endothelial cells, it is essential for R. parkeri to block autophagy in macrophages and to colonize mice because of its ability to promote autophagy evasion in immune cells. Our results indicate that OmpB acts as a protective shield to obstruct autophagy recognition, thereby revealing a distinctive bacterial mechanism to evade antimicrobial autophagy.
Conflict of interest statement
COMPETING INTEREST
The authors declare no competing interest.
Figures