A complete data processing workflow for cryo-ET and subtomogram averaging
- PMID: 31611690
- PMCID: PMC6858567
- DOI: 10.1038/s41592-019-0591-8
A complete data processing workflow for cryo-ET and subtomogram averaging
Abstract
Electron cryotomography is currently the only method capable of visualizing cells in three dimensions at nanometer resolutions. While modern instruments produce massive amounts of tomography data containing extremely rich structural information, data processing is very labor intensive and the results are often limited by the skills of the personnel rather than the data. We present an integrated workflow that covers the entire tomography data processing pipeline, from automated tilt series alignment to subnanometer resolution subtomogram averaging. Resolution enhancement is made possible through the use of per-particle per-tilt contrast transfer function correction and alignment. The workflow greatly reduces human bias, increases throughput and more closely approaches data-limited resolution for subtomogram averaging in both purified macromolecules and cells.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures



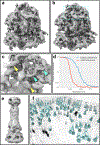
References
-
- Asano S, Engel BD & Baumeister W In Situ Cryo-Electron Tomography: A Post-Reductionist Approach to Structural Biology. J. Mol. Biol 428, 332–343 (2016). - PubMed
-
- Kremer JR, Mastronarde DN & McIntosh JR Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol 116, 71–76 (1996). - PubMed
-
- Amat F et al. Markov random field based automatic image alignment for electron tomography. J. Struct. Biol 161, 260–75 (2008). - PubMed
-
- Hrabe T et al. PyTom: A python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J. Struct. Biol 178, 177–188 (2012). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

