Predicting HLA class II antigen presentation through integrated deep learning
- PMID: 31611695
- PMCID: PMC7075463
- DOI: 10.1038/s41587-019-0280-2
Predicting HLA class II antigen presentation through integrated deep learning
Abstract
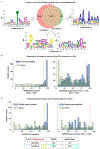
Accurate prediction of antigen presentation by human leukocyte antigen (HLA) class II molecules would be valuable for vaccine development and cancer immunotherapies. Current computational methods trained on in vitro binding data are limited by insufficient training data and algorithmic constraints. Here we describe MARIA (major histocompatibility complex analysis with recurrent integrated architecture; https://maria.stanford.edu/ ), a multimodal recurrent neural network for predicting the likelihood of antigen presentation from a gene of interest in the context of specific HLA class II alleles. In addition to in vitro binding measurements, MARIA is trained on peptide HLA ligand sequences identified by mass spectrometry, expression levels of antigen genes and protease cleavage signatures. Because it leverages these diverse training data and our improved machine learning framework, MARIA (area under the curve = 0.89-0.92) outperformed existing methods in validation datasets. Across independent cancer neoantigen studies, peptides with high MARIA scores are more likely to elicit strong CD4+ T cell responses. MARIA allows identification of immunogenic epitopes in diverse cancers and autoimmune disease.
Conflict of interest statement
Competing interests
A.A.A. declares the following competing interests: stock or other ownership (CiberMed and Forty Seven); honoraria (Janssen Oncology); consulting or advisory roles (Celgene, Roche/Genentech and Gilead Sciences); research funding (Celgene); patents, royalties or other intellectual property (patent filings on immune deconvolution and circulating tumor DNA detection assigned to Stanford University); and travel, accommodations or expenses (Roche and Gilead Sciences). R.B.A. declares the following competing interests: stock or other ownership (Personalis); consulting or advisory role (Pfizer, Youscript, 23andme and WithHealth); patents, royalties or other intellectual property (royalties for patents related to genome sequencing).
Figures



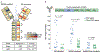

Comment in
-
Improved MHC II epitope prediction - a step towards personalized medicine.Nat Rev Clin Oncol. 2020 Feb;17(2):71-72. doi: 10.1038/s41571-019-0315-0. Nat Rev Clin Oncol. 2020. PMID: 31836878 Free PMC article.
References
-
- Neefjes J, Jongsma ML, Paul P & Bakke O Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol 11, 823–836 (2011). - PubMed
-
- Schreiber RD, Old LJ & Smyth MJ Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

