Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood-brain barrier dysfunction module
- PMID: 31611708
- PMCID: PMC6858546
- DOI: 10.1038/s41593-019-0497-x
Profiling the mouse brain endothelial transcriptome in health and disease models reveals a core blood-brain barrier dysfunction module
Abstract
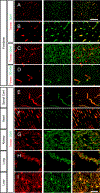
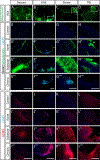
Blood vessels in the CNS form a specialized and critical structure, the blood-brain barrier (BBB). We present a resource to understand the molecular mechanisms that regulate BBB function in health and dysfunction during disease. Using endothelial cell enrichment and RNA sequencing, we analyzed the gene expression of endothelial cells in mice, comparing brain endothelial cells with peripheral endothelial cells. We also assessed the regulation of CNS endothelial gene expression in models of stroke, multiple sclerosis, traumatic brain injury and seizure, each having profound BBB disruption. We found that although each is caused by a distinct trigger, they exhibit strikingly similar endothelial gene expression changes during BBB disruption, comprising a core BBB dysfunction module that shifts the CNS endothelial cells into a peripheral endothelial cell-like state. The identification of a common pathway for BBB dysfunction suggests that targeting therapeutic agents to limit it may be effective across multiple neurological disorders.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures




References
-
- Daneman R. The blood-brain barrier in health and disease. Ann Neurol 72, 648–672 (2012). - PubMed
-
- Zlokovic BV. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron 57, 178–201 (2008). - PubMed
-
- Rubin LL & Staddon JM. The cell biology of the blood-brain barrier. Annu Rev Neurosci 22, 11–28 (1999). - PubMed
-
- Janzer RC & Raff MC. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature 325, 253–257 (1987). - PubMed
-
- Stewart PA & Wiley MJ. Developing nervous tissue induces formation of blood-brain barrier characteristics in invading endothelial cells: a study using quail--chick transplantation chimeras. Dev Biol 84, 183–192 (1981). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

