An Overview of Genes From Cyberlindnera americana, a Symbiont Yeast Isolated From the Gut of the Bark Beetle Dendroctonus rhizophagus (Curculionidae: Scolytinae), Involved in the Detoxification Process Using Genome and Transcriptome Data
- PMID: 31611850
- PMCID: PMC6777644
- DOI: 10.3389/fmicb.2019.02180
An Overview of Genes From Cyberlindnera americana, a Symbiont Yeast Isolated From the Gut of the Bark Beetle Dendroctonus rhizophagus (Curculionidae: Scolytinae), Involved in the Detoxification Process Using Genome and Transcriptome Data
Abstract
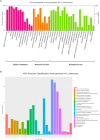
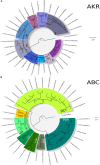
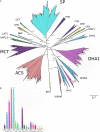
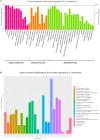
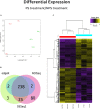
Bark beetles from Dendroctonus genus promote ecological succession and nutrient cycling in coniferous forests. However, they can trigger outbreaks leading to important economic losses in the forest industry. Conifers have evolved resistance mechanisms that can be toxic to insects but at the same time, bark beetles are capable of overcoming tree barriers and colonize these habitats. In this sense, symbiont yeasts present in the gut of bark beetles have been suggested to play a role in the detoxification process of tree defensive chemicals. In the present study, genes related to this process were identified and their response to a terpene highly toxic to bark beetles and their symbionts was analyzed in the Cyberlindnera americana yeast. The genome and transcriptome of C. americana (ChDrAdgY46) isolated from the gut of Dendroctonus rhizophagus were presented. Genome analysis identified 5752 protein-coding genes and diverse gene families associated with the detoxification process. The most abundant belonged to the Aldo-Keto Reductase Superfamily, ATP-binding cassette Superfamily, and the Major Facilitator Superfamily transporters. The transcriptome analysis of non-α-pinene stimulated and α-pinene stimulated yeasts showed a significant expression of genes belonging to these families. The activities demonstrated by the genes identified as Aryl-alcohol dehydrogenase and ABC transporter under (+)-α-pinene suggest that they are responsible, that C. americana is a dominant symbiont that resists high amounts of monoterpenes inside the gut of bark beetles.
Keywords: MDR transporters; bark beetles; detoxification; genome; symbiont yeast; transcriptome; α-pinene.
Copyright © 2019 Soto-Robles, Torres-Banda, Rivera-Orduña, Curiel-Quesada, Hidalgo-Lara and Zúñiga.
Figures

References
-
- Adams A. S., Aylward F. O., Adams S. M., Erbilgin N., Aukema B. H., Currie C. R., et al. (2013). Mountain pine beetles colonizing historical and naive host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl. Environ. Microb. 79 3468–3475. 10.1128/AEM.00068-13 - DOI - PMC - PubMed
-
- Adams A. S., Currie C. R., Cardoza Y., Klepzig K. D., Raffa K. F. (2009). Effects of symbiotic bacteria and tree chemistry on the growth and reproduction of bark beetle fungal symbionts. Can. J. For. Res. 39 1133–1147. 10.1139/X09-034 - DOI
LinkOut - more resources
Full Text Sources

