A Novel DNA Methylation-Based Signature Can Predict the Responses of MGMT Promoter Unmethylated Glioblastomas to Temozolomide
- PMID: 31611911
- PMCID: PMC6776832
- DOI: 10.3389/fgene.2019.00910
A Novel DNA Methylation-Based Signature Can Predict the Responses of MGMT Promoter Unmethylated Glioblastomas to Temozolomide
Abstract
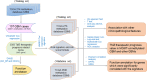
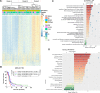
Glioblastoma (GBM) is the most malignant glioma, with a median overall survival (OS) of 14-16 months. Temozolomide (TMZ) is the first-line chemotherapy drug for glioma, but whether TMZ should be withheld from patients with GBMs that lack O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation is still under debate. DNA methylation profiling holds great promise for further stratifying the responses of MGMT promoter unmethylated GBMs to TMZ. In this study, we studied 147 TMZ-treated MGMT promoter unmethylated GBM, whose methylation information was obtained from the HumanMethylation27 (HM-27K) BeadChips (n = 107) and the HumanMethylation450 (HM-450K) BeadChips (n = 40) for training and validation, respectively. In the training set, we performed univariate Cox regression and identified that 3,565 CpGs were significantly associated with the OS of the TMZ-treated MGMT promoter unmethylated GBMs. Functional analysis indicated that the genes corresponding to these CpGs were enriched in the biological processes or pathways of mitochondrial translation, cell cycle, and DNA repair. Based on these CpGs, we developed a 31-CpGs methylation signature utilizing the least absolute shrinkage and selection operator (LASSO) Cox regression algorithm. In both training and validation datasets, the signature identified the TMZ-sensitive GBMs in the MGMT promoter unmethylated GBMs, and only the patients in the low-risk group appear to benefit from the TMZ treatment. Furthermore, these identified TMZ-sensitive MGMT promoter unmethylated GBMs have a similar OS when compared with the MGMT promoter methylated GBMs after TMZ treatment in both two datasets. Multivariate Cox regression demonstrated the independent prognostic value of the signature in TMZ-treated MGMT promoter unmethylated GBMs. Moreover, we also noticed that the hallmark of epithelial-mesenchymal transition, ECM related biological processes and pathways were highly enriched in the MGMT unmethylated GBMs with the high-risk score, indicating that enhanced ECM activities could be involved in the TMZ-resistance of GBM. In conclusion, our findings promote our understanding of the roles of DNA methylation in MGMT umethylated GBMs and offer a very promising TMZ-sensitivity predictive signature for these GBMs that could be tested prospectively.
Keywords: DNA methylation; MGMT; glioblastoma; signature; temozolomide.
Copyright © 2019 Chai, Chang, Wang, Zhang, Li, Huang, Wu, Liu and Wang.
Figures



References
-
- Chai R. C., Wang N., Chang Y. Z., Zhang K. N., Li J. J., Niu J. J., et al. (2019. b). Systematically profiling the expression of eIF3 subunits in glioma reveals the expression of eIF3i has prognostic value in IDH-mutant lower grade glioma. Cancer Cell Int. 19, 155. 10.1186/s12935-019-0867-1 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

