An improved platform for functional assessment of large protein libraries in mammalian cells
- PMID: 31612958
- PMCID: PMC7145622
- DOI: 10.1093/nar/gkz910
An improved platform for functional assessment of large protein libraries in mammalian cells
Abstract
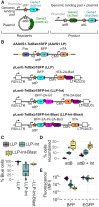
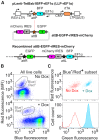
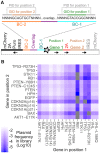
Multiplex genetic assays can simultaneously test thousands of genetic variants for a property of interest. However, limitations of existing multiplex assay methods in cultured mammalian cells hinder the breadth, speed and scale of these experiments. Here, we describe a series of improvements that greatly enhance the capabilities of a Bxb1 recombinase-based landing pad system for conducting different types of multiplex genetic assays in various mammalian cell lines. We incorporate the landing pad into a lentiviral vector, easing the process of generating new landing pad cell lines. We also develop several new landing pad versions, including one where the Bxb1 recombinase is expressed from the landing pad itself, improving recombination efficiency more than 2-fold and permitting rapid prototyping of transgenic constructs. Other versions incorporate positive and negative selection markers that enable drug-based enrichment of recombinant cells, enabling the use of larger libraries and reducing costs. A version with dual convergent promoters allows enrichment of recombinant cells independent of transgene expression, permitting the assessment of libraries of transgenes that perturb cell growth and survival. Lastly, we demonstrate these improvements by assessing the effects of a combinatorial library of oncogenes and tumor suppressors on cell growth. Collectively, these advancements make multiplex genetic assays in diverse cultured cell lines easier, cheaper and more effective, facilitating future studies probing how proteins impact cell function, using transgenic variant libraries tested individually or in combination.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Forsyth C.M., Juan V., Akamatsu Y., DuBridge R.B., Doan M., Ivanov A.V., Ma Z., Polakoff D., Razo J., Wilson K. et al. .. Deep mutational scanning of an antibody against epidermal growth factor receptor using mammalian cell display and massively parallel pyrosequencing. MAbs. 2013; 5:523–532. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

