Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self-organized fate patterning in human gastruloids
- PMID: 31613879
- PMCID: PMC6814242
- DOI: 10.1371/journal.pbio.3000498
Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self-organized fate patterning in human gastruloids
Abstract
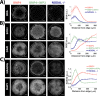
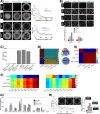
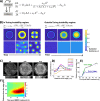
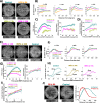
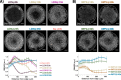
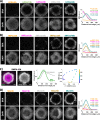
During gastrulation, the pluripotent epiblast self-organizes into the 3 germ layers-endoderm, mesoderm and ectoderm, which eventually form the entire embryo. Decades of research in the mouse embryo have revealed that a signaling cascade involving the Bone Morphogenic Protein (BMP), WNT, and NODAL pathways is necessary for gastrulation. In vivo, WNT and NODAL ligands are expressed near the site of gastrulation in the posterior of the embryo, and knockout of these ligands leads to a failure to gastrulate. These data have led to the prevailing view that a signaling gradient in WNT and NODAL underlies patterning during gastrulation; however, the activities of these pathways in space and time have never been directly observed. In this study, we quantify BMP, WNT, and NODAL signaling dynamics in an in vitro model of human gastrulation. Our data suggest that BMP signaling initiates waves of WNT and NODAL signaling activity that move toward the colony center at a constant rate. Using a simple mathematical model, we show that this wave-like behavior is inconsistent with a reaction-diffusion-based Turing system, indicating that there is no stable signaling gradient of WNT/NODAL. Instead, the final signaling state is homogeneous, and spatial differences arise only from boundary effects. We further show that the durations of WNT and NODAL signaling control mesoderm differentiation, while the duration of BMP signaling controls differentiation of CDX2-positive extra-embryonic cells. The identity of these extra-embryonic cells has been controversial, and we use RNA sequencing (RNA-seq) to obtain their transcriptomes and show that they closely resemble human trophoblast cells in vivo. The domain of BMP signaling is identical to the domain of differentiation of these trophoblast-like cells; however, neither WNT nor NODAL forms a spatial pattern that maps directly to the mesodermal region, suggesting that mesoderm differentiation is controlled dynamically by the combinatorial effect of multiple signals. We synthesize our data into a mathematical model that accurately recapitulates signaling dynamics and predicts cell fate patterning upon chemical and physical perturbations. Taken together, our study shows that the dynamics of signaling events in the BMP, WNT, and NODAL cascade in the absence of a stable signaling gradient control fate patterning of human gastruloids.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

