Drug screens of NGLY1 deficiency in worm and fly models reveal catecholamine, NRF2 and anti-inflammatory-pathway activation as potential clinical approaches
- PMID: 31615832
- PMCID: PMC6899034
- DOI: 10.1242/dmm.040576
Drug screens of NGLY1 deficiency in worm and fly models reveal catecholamine, NRF2 and anti-inflammatory-pathway activation as potential clinical approaches
Abstract
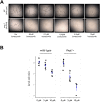
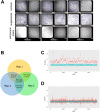
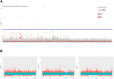
N-glycanase 1 (NGLY1) deficiency is an ultra-rare and complex monogenic glycosylation disorder that affects fewer than 40 patients globally. NGLY1 deficiency has been studied in model organisms such as yeast, worms, flies and mice. Proteasomal and mitochondrial homeostasis gene networks are controlled by the evolutionarily conserved transcriptional regulator NRF1, whose activity requires deglycosylation by NGLY1. Hypersensitivity to the proteasome inhibitor bortezomib is a common phenotype observed in whole-animal and cellular models of NGLY1 deficiency. Here, we describe unbiased phenotypic drug screens to identify FDA-approved drugs that are generally recognized as safe natural products, and novel chemical entities, that rescue growth and development of NGLY1-deficient worm and fly larvae treated with a toxic dose of bortezomib. We used image-based larval size and number assays for use in screens of a 2560-member drug-repurposing library and a 20,240-member lead-discovery library. A total of 91 validated hit compounds from primary invertebrate screens were tested in a human cell line in an NRF2 activity assay. NRF2 is a transcriptional regulator that regulates cellular redox homeostasis, and it can compensate for loss of NRF1. Plant-based polyphenols make up the largest class of hit compounds and NRF2 inducers. Catecholamines and catecholamine receptor activators make up the second largest class of hits. Steroidal and non-steroidal anti-inflammatory drugs make up the third largest class. Only one compound was active in all assays and species: the atypical antipsychotic and dopamine receptor agonist aripiprazole. Worm and fly models of NGLY1 deficiency validate therapeutic rationales for activation of NRF2 and anti-inflammatory pathways based on results in mice and human cell models, and suggest a novel therapeutic rationale for boosting catecholamine levels and/or signaling in the brain.
Keywords: Aripiprazole; Congenital disorder of deglycosylation; Disease model; N-glycanase 1 deficiency; NGLY1; Pngl; SKN-1.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
-
- Costa M. D. C., Ashraf N. S., Fischer S., Yang Y., Schapka E., Joshi G., McQuade T. J., Dharia R. M., Dulchavsky M., Ouyang M. et al. (2016). Unbiased screen identifies aripiprazole as a modulator of abundance of the polyglutamine disease protein, ataxin-3. Brain 139, 2891-2908. 10.1093/brain/aww228 - DOI - PMC - PubMed
-
- Dziekan J. M., Yu H., Chen D., Dai L., Wirjanata G., Larsson A., Prabhu N., Sobota R. M., Bozdech Z. and Nordlund P. (2019). Identifying purine nucleoside phosphorylase as the target of quinine using cellular thermal shift assay. Sci. Transl. Med. 11, eaau3174 10.1126/scitranslmed.aau3174 - DOI - PubMed
-
- Enns G. M., Shashi V., Bainbridge M., Gambello M. J., Zahir F. R., Bast T., Crimian R., Schoch K., Platt J., Cox R. et al. (2014). Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genet. Med. 16, 751-758. 10.1038/gim.2014.22 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

