Genome-wide association mapping of date palm fruit traits
- PMID: 31615981
- PMCID: PMC6794320
- DOI: 10.1038/s41467-019-12604-9
Genome-wide association mapping of date palm fruit traits
Abstract
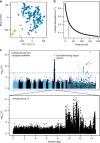
Date palms (Phoenix dactylifera) are an important fruit crop of arid regions of the Middle East and North Africa. Despite its importance, few genomic resources exist for date palms, hampering evolutionary genomic studies of this perennial species. Here we report an improved long-read genome assembly for P. dactylifera that is 772.3 Mb in length, with contig N50 of 897.2 Kb, and use this to perform genome-wide association studies (GWAS) of the sex determining region and 21 fruit traits. We find a fruit color GWAS at the R2R3-MYB transcription factor VIRESCENS gene and identify functional alleles that include a retrotransposon insertion and start codon mutation. We also find a GWAS peak for sugar composition spanning deletion polymorphisms in multiple linked invertase genes. MYB transcription factors and invertase are implicated in fruit color and sugar composition in other crops, demonstrating the importance of parallel evolution in the evolutionary diversification of domesticated species.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Barrow S. A monograph of Phoenix L. (Palmae: Coryphoideae) Kew Bull. 1998;53:513–575. doi: 10.2307/4110478. - DOI
-
- Tengberg M. Beginnings and early history of date palm garden cultivation in the Middle East. J. Arid Environ. 2012;86:139–147. doi: 10.1016/j.jaridenv.2011.11.022. - DOI
-
- Flowers Jonathan M., Hazzouri Khaled M., Gros-Balthazard Muriel, Mo Ziyi, Koutroumpa Konstantina, Perrakis Andreas, Ferrand Sylvie, Khierallah Hussam S. M., Fuller Dorian Q., Aberlenc Frederique, Fournaraki Christini, Purugganan Michael D. Cross-species hybridization and the origin of North African date palms. Proceedings of the National Academy of Sciences. 2019;116(5):1651–1658. doi: 10.1073/pnas.1817453116. - DOI - PMC - PubMed
-
- Chao CT, Krueger RR. The date palm (Phoenix dactylifera l.): overview of biology, uses and cultivation. HortScience. 2007;42:1077–1082. doi: 10.21273/HORTSCI.42.5.1077. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

