Site-specific N-glycosylation of HeLa cell glycoproteins
- PMID: 31616032
- PMCID: PMC6794373
- DOI: 10.1038/s41598-019-51428-x
Site-specific N-glycosylation of HeLa cell glycoproteins
Abstract
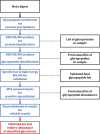
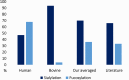
We have characterized site-specific N-glycosylation of the HeLa cell line glycoproteins, using a complex workflow based on high and low energy tandem mass spectrometry of glycopeptides. The objective was to obtain highly reliable data on common glycoforms, so rigorous data evaluation was performed. The analysis revealed the presence of a high amount of bovine serum contaminants originating from the cell culture media - nearly 50% of all glycans were of bovine origin. Unaccounted, the presence of bovine serum components causes major bias in the human cellular glycosylation pattern; as is shown when literature results using released glycan analysis are compared. We have reliably identified 43 (human) glycoproteins, 69 N-glycosylation sites, and 178 glycoforms. HeLa glycoproteins were found to be highly (68.7%) fucosylated. A medium degree of sialylation was observed, on average 46.8% of possible sialylation sites were occupied. High-mannose sugars were expressed in large amounts, as expected in the case of a cancer cell line. Glycosylation in HeLa cells is highly variable. It is markedly different not only on various proteins but also at the different glycosylation sites of the same protein. Our method enabled the detailed characterization of site-specific N-glycosylation of several glycoproteins expressed in HeLa cell line.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Thiede B, et al. High resolution quantitative proteomics of HeLa cells protein species using stable isotope labeling with amino acids in cell culture(SILAC), two-dimensional gel electrophoresis(2DE) and nano-liquid chromatograpohy coupled to an LTQ-OrbitrapMass spectrometer. Mol Cell Proteomics. 2013;12:529–538. doi: 10.1074/mcp.M112.019372. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

