Transcriptomic Identification and Biochemical Characterization of HmpA, a Nitric Oxide Dioxygenase, Essential for Pathogenesis of Vibrio vulnificus
- PMID: 31616401
- PMCID: PMC6768983
- DOI: 10.3389/fmicb.2019.02208
Transcriptomic Identification and Biochemical Characterization of HmpA, a Nitric Oxide Dioxygenase, Essential for Pathogenesis of Vibrio vulnificus
Abstract
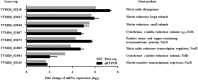
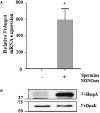
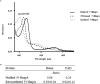
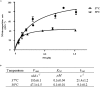
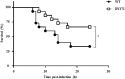
Nitric oxide (NO) and its derivatives are important effectors of host innate immunity, disrupting cellular function of infecting pathogens. Transcriptome analysis of Vibrio vulnificus, an opportunistic human pathogen, identified a set of genes induced upon exposure to NO. Among them, VvhmpA (V. vulnificus hmpA), encoding a multidomain NO dioxygenase, was the most greatly induced upon exposure to NO and was thus further characterized. Absorption spectra demonstrated that VvHmpA is a heme protein in which the heme iron can exist in either reduced, NO-bound, or oxidized state. Biochemical studies revealed that VvHmpA is a flavohemoglobin containing equimolar amounts of heme and FAD as cofactors. The K M and k cat values of VvHmpA for NO at 37°C, the temperature encountered by V. vulnificus in the host, were greater than those at 30°C, indicating that VvHmpA detoxifies high levels of NO effectively during infection. Compared with the wild type, the VvhmpA mutant exhibited a lower NO-decomposition activity and impaired growth in the presence of NO in vitro. Also, the cytotoxicity and survival of the VvhmpA mutant infecting the NO-producing murine macrophage cells were lower than those of the wild type. Furthermore, the mouse lethality of the VvhmpA mutant was reduced compared to that of the parental wild type. The combined results revealed that VvHmpA is a potent virulence factor that is induced upon exposure to NO and important for the survival and pathogenesis of V. vulnificus during infection.
Keywords: Vibrio vulnificus; flavohemoglobins; gene expression profiling; microbiology; nitric oxide dioxygenase; virulence factors.
Copyright © 2019 Kim, Na, Kim, Kim, Jung, Cao, Han, Bang, Yoo, Ha and Choi.
Figures



Similar articles
-
A Nitric Oxide-Responsive Transcriptional Regulator NsrR Cooperates With Lrp and CRP to Tightly Control the hmpA Gene in Vibrio vulnificus.Front Microbiol. 2021 May 21;12:681196. doi: 10.3389/fmicb.2021.681196. eCollection 2021. Front Microbiol. 2021. PMID: 34093504 Free PMC article.
-
Vibrio vulnificus RtxA1 Toxin Expression Upon Contact With Host Cells Is RpoS-Dependent.Front Cell Infect Microbiol. 2018 Mar 15;8:70. doi: 10.3389/fcimb.2018.00070. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29600196 Free PMC article.
-
Phospholipase A as a potent virulence factor of Vibrio vulnificus.Int J Mol Med. 2007 Dec;20(6):913-8. Int J Mol Med. 2007. PMID: 17982702
-
Identification and Validation of an Antivirulence Agent Targeting HlyU-Regulated Virulence in Vibrio vulnificus.Front Cell Infect Microbiol. 2018 May 11;8:152. doi: 10.3389/fcimb.2018.00152. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29868508 Free PMC article.
-
Quorum sensing-dependent metalloprotease VvpE is important in the virulence of Vibrio vulnificus to invertebrates.Microb Pathog. 2014 Jun-Jul;71-72:8-14. doi: 10.1016/j.micpath.2014.04.001. Epub 2014 Apr 21. Microb Pathog. 2014. PMID: 24769338
Cited by
-
CarRS Two-Component System Essential for Polymyxin B Resistance of Vibrio vulnificus Responds to Multiple Host Environmental Signals.Microbiol Spectr. 2023 Aug 17;11(4):e0030523. doi: 10.1128/spectrum.00305-23. Epub 2023 Jun 8. Microbiol Spectr. 2023. PMID: 37289068 Free PMC article.
-
Exploring the Role of the NO-Detoxifying Enzyme HmpA in the Evolution of Domesticated Alfalfa Rhizobia.Microb Ecol. 2022 Feb;83(2):501-505. doi: 10.1007/s00248-021-01761-4. Epub 2021 May 8. Microb Ecol. 2022. PMID: 33966095
-
A Nitric Oxide-Responsive Transcriptional Regulator NsrR Cooperates With Lrp and CRP to Tightly Control the hmpA Gene in Vibrio vulnificus.Front Microbiol. 2021 May 21;12:681196. doi: 10.3389/fmicb.2021.681196. eCollection 2021. Front Microbiol. 2021. PMID: 34093504 Free PMC article.
-
A Master Regulator BrpR Coordinates the Expression of Multiple Loci for Robust Biofilm and Rugose Colony Development in Vibrio vulnificus.Front Microbiol. 2021 Jun 25;12:679854. doi: 10.3389/fmicb.2021.679854. eCollection 2021. Front Microbiol. 2021. PMID: 34248894 Free PMC article.
-
The Role of TSC1 in the Macrophages Against Vibrio vulnificus Infection.Front Cell Infect Microbiol. 2021 Jan 27;10:596609. doi: 10.3389/fcimb.2020.596609. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33585271 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

