MMOD-induced structural changes of hydroxylase in soluble methane monooxygenase
- PMID: 31616787
- PMCID: PMC6774732
- DOI: 10.1126/sciadv.aax0059
MMOD-induced structural changes of hydroxylase in soluble methane monooxygenase
Abstract
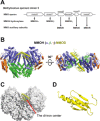
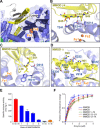
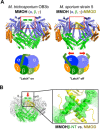
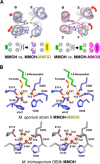
Soluble methane monooxygenase in methanotrophs converts methane to methanol under ambient conditions. The maximum catalytic activity of hydroxylase (MMOH) is achieved through the interplay of its regulatory protein (MMOB) and reductase. An additional auxiliary protein, MMOD, functions as an inhibitor of MMOH; however, its inhibitory mechanism remains unknown. Here, we report the crystal structure of the MMOH-MMOD complex from Methylosinus sporium strain 5 (2.6 Å). Its structure illustrates that MMOD associates with the canyon region of MMOH where MMOB binds. Although MMOD and MMOB recognize the same binding site, each binding component triggers different conformational changes toward MMOH, which then respectively lead to the inhibition and activation of MMOH. Particularly, MMOD binding perturbs the di-iron geometry by inducing two major MMOH conformational changes, i.e., MMOH β subunit disorganization and subsequent His147 dissociation with Fe1 coordination. Furthermore, 1,6-hexanediol, a mimic of the products of sMMO, reveals the substrate access route.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures

References
-
- Higgins I. J., Best D. J., Hammond R. C., New findings in methane-utilizing bacteria highlight their importance in the biosphere and their commercial potential. Nature 286, 561–564 (1980). - PubMed
-
- Wang V. C.-C., Maji S., Chen P. P.-Y., Lee H. K., Yu S. S.-F., Chan S. I., Alkane oxidation: Methane monooxygenases, related enzymes, and their biomimetics. Chem. Rev. 117, 8574–8621 (2017). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

