O-glycosylation on cerebrospinal fluid and plasma apolipoprotein E differs in the lipid-binding domain
- PMID: 31616924
- PMCID: PMC7335482
- DOI: 10.1093/glycob/cwz084
O-glycosylation on cerebrospinal fluid and plasma apolipoprotein E differs in the lipid-binding domain
Abstract
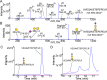
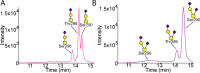
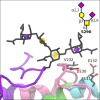
The O-glycoprotein apolipoprotein E (APOE), the strongest genetic risk factor for Alzheimer's disease, associates with lipoproteins. Cerebrospinal fluid (CSF) APOE binds only high-density lipoproteins (HDLs), while plasma APOE attaches to lipoproteins of diverse sizes with binding fine-tuned by the C-terminal loop. To better understand the O-glycosylation on this critical molecule and differences across tissues, we analyzed the O-glycosylation on APOE isolated from the plasma and CSF of aged individuals. Detailed LC-MS/MS analyses allowed the identification of the glycosite and the attached glycan and site occupancy for all detectable glycosites on APOE and further three-dimensional modeling of physiological glycoforms of APOE. APOE is O-glycosylated at several sites: Thr8, Thr18, Thr194, Ser197, Thr289, Ser290 and Ser296. Plasma APOE held more abundant (20.5%) N-terminal (Thr8) sialylated core 1 (Neu5Acα2-3Galβ1-3GalNAcα1-) glycosylation compared to CSF APOE (0.1%). APOE was hinge domain glycosylated (Thr194 and Ser197) in both CSF (27.3%) and plasma (10.3%). CSF APOE held almost 10-fold more abundant C-terminal (Thr289, Ser290 and Ser296) glycosylation (36.8% of CSF peptide283-299 was glycosylated, 3.8% of plasma peptide283-299), with sialylated and disialylated (Neu5Acα2-3Galβ1-3(Neu5Acα2-6) GalNAcα1-) core 1 structures. Modeling suggested that C-terminal glycosylation, particularly the branched disialylated structure, could interact across domains including the receptor-binding domain. These data, although limited by sample size, suggest that there are tissue-specific APOE glycoforms. Sialylated glycans, previously shown to improve HDL binding, are more abundant on the lipid-binding domain of CSF APOE and reduced in plasma APOE. This indicates that APOE glycosylation may be implicated in lipoprotein-binding flexibility.
Keywords: APOE; Alzheimer’s disease; glycoproteomics; lipoprotein binding; tissue-specific glycosylation.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



References
-
- Beisiegel U. 1998. Lipoprotein metabolism. Eur Heart Journal. 19(Suppl A):A20–A23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

