Carbon/nitrogen homeostasis control in cyanobacteria
- PMID: 31617886
- PMCID: PMC8042125
- DOI: 10.1093/femsre/fuz025
Carbon/nitrogen homeostasis control in cyanobacteria
Abstract
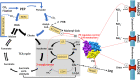
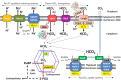
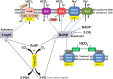
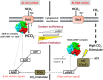
Carbon/nitrogen (C/N) balance sensing is a key requirement for the maintenance of cellular homeostasis. Therefore, cyanobacteria have evolved a sophisticated signal transduction network targeting the metabolite 2-oxoglutarate (2-OG), the carbon skeleton for nitrogen assimilation. It serves as a status reporter for the cellular C/N balance that is sensed by transcription factors NtcA and NdhR and the versatile PII-signaling protein. The PII protein acts as a multitasking signal-integrating regulator, combining the 2-OG signal with the energy state of the cell through adenyl-nucleotide binding. Depending on these integrated signals, PII orchestrates metabolic activities in response to environmental changes through binding to various targets. In addition to 2-OG, other status reporter metabolites have recently been discovered, mainly indicating the carbon status of the cells. One of them is cAMP, which is sensed by the PII-like protein SbtB. The present review focuses, with a main emphasis on unicellular model strains Synechoccus elongatus and Synechocystis sp. PCC 6803, on the physiological framework of these complex regulatory loops, the tight linkage to metabolism and the molecular mechanisms governing the signaling processes.
Keywords: 2-oxoglutarate; CO2 metabolism; arginine synthesis; energy sensing; nitrogen assimilation; protein-serine-phosphorylation.
© FEMS 2019.
Figures



References
-
- Allen MM, Smith AJ. Nitrogen chlorosis in blue-green algae. Arch Microbiol. 1969;69:114–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

