Clonality analysis of pulmonary tumors by genome-wide copy number profiling
- PMID: 31618260
- PMCID: PMC6795528
- DOI: 10.1371/journal.pone.0223827
Clonality analysis of pulmonary tumors by genome-wide copy number profiling
Erratum in
-
Correction: Clonality analysis of pulmonary tumors by genome-wide copy number profiling.PLoS One. 2019 Nov 19;14(11):e0225733. doi: 10.1371/journal.pone.0225733. eCollection 2019. PLoS One. 2019. PMID: 31743370 Free PMC article.
Abstract
Multiple tumors in patients are frequently diagnosed, either synchronous or metachronous. The distinction between a second primary and a metastasis is important for treatment. Chromosomal DNA copy number aberrations (CNA) patterns are highly unique to specific tumors. The aim of this study was to assess genome-wide CNA-patterns as method to identify clonally related tumors in a prospective cohort of patients with synchronous or metachronous tumors, with at least one intrapulmonary tumor. In total, 139 tumor pairs from 90 patients were examined: 35 synchronous and 104 metachronous pairs. Results of CNA were compared to histological type, clinicopathological methods (Martini-Melamed-classification (MM) and ACCP-2013-criteria), and, if available, EGFR- and KRAS-mutation analysis. CNA-results were clonal in 74 pairs (53%), non-clonal in 33 pairs (24%), and inconclusive in 32 pairs (23%). Histological similarity was found in 130 pairs (94%). Concordance between histology and conclusive CNA-results was 69% (74 of 107 pairs: 72 clonal and two non-clonal). In 31 of 103 pairs with similar histology, genetics revealed non-clonality. In two out of four pairs with non-matching histology, genetics revealed clonality. The subgroups of synchronous and metachronous pairs showed similar outcome for the comparison of histological versus CNA-results. MM-classification and ACCP-2013-criteria, applicable on 34 pairs, and CNA-results were concordant in 50% and 62% respectively. Concordance between mutation matching and conclusive CNA-results was 89% (8 of 9 pairs: six clonal and two non-clonal). Interestingly, in one patient both tumors had the same KRAS mutation, but the CNA result was non-clonal. In conclusion, although some concordance between histological comparison and CNA profiling is present, arguments exist to prefer extensive molecular testing to determine whether a second tumor is a metastasis or a second primary.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
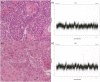
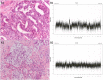
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

