Genetic and Lineage Classification of Glioma-Initiating Cells Identifies a Clinically Relevant Glioblastoma Model
- PMID: 31618934
- PMCID: PMC6826962
- DOI: 10.3390/cancers11101564
Genetic and Lineage Classification of Glioma-Initiating Cells Identifies a Clinically Relevant Glioblastoma Model
Abstract
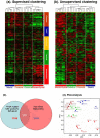
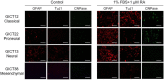
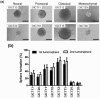
The Cancer Genome Atlas (TCGA) project described a robust gene expression-based molecular classification of glioblastoma (GBM), but the functional and biological significance of the subclasses has not been determined. The present comprehensive analysis of 25 glioma-initiating cell (GIC) lines classifies GIC lines into four subtypes (classical, mesenchymal, proneural, and neural) that are closely related to the TCGA GBM subclasses and display distinct lineage characteristics and differentiation behavior that recapitulate neural development. More importantly, the GIC subtypes exhibit distinct biological phenotypes in relation to self-renewal capacity, proliferation, invasiveness, and angiogenic potential in vitro and in vivo. In addition, the GIC subtypes exhibit divergent patterns of signaling pathway activation and deactivation of the Wnt, Notch, and TGF-β pathways. These results will improve drug discovery targeting certain genetic mutation in glioblastoma and improve the development of precision medicine.
Keywords: The Cancer Genome Atlas; glioblastoma; glioma initiating cell; molecular classification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

