Divergent effects of translation termination factor eRF3A and nonsense-mediated mRNA decay factor UPF1 on the expression of uORF carrying mRNAs and ribosome protein genes
- PMID: 31619139
- PMCID: PMC6973328
- DOI: 10.1080/15476286.2019.1674595
Divergent effects of translation termination factor eRF3A and nonsense-mediated mRNA decay factor UPF1 on the expression of uORF carrying mRNAs and ribosome protein genes
Abstract
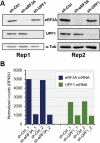
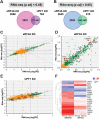
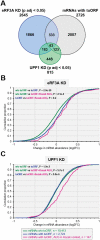
In addition to its role in translation termination, eRF3A has been implicated in the nonsense-mediated mRNA decay (NMD) pathway through its interaction with UPF1. NMD is a RNA quality control mechanism, which detects and degrades aberrant mRNAs as well as some normal transcripts including those that harbour upstream open reading frames in their 5' leader sequence. In this study, we used RNA-sequencing and ribosome profiling to perform a genome wide analysis of the effect of either eRF3A or UPF1 depletion in human cells. Our bioinformatics analyses allow to delineate the features of the transcripts controlled by eRF3A and UPF1 and to compare the effect of each of these factors on gene expression. We find that eRF3A and UPF1 have very different impacts on the human transcriptome, less than 250 transcripts being targeted by both factors. We show that eRF3A depletion globally derepresses the expression of mRNAs containing translated uORFs while UPF1 knockdown derepresses only the mRNAs harbouring uORFs with an AUG codon in an optimal context for translation initiation. Finally, we also find that eRF3A and UPF1 have opposite effects on ribosome protein gene expression. Together, our results provide important elements for understanding the impact of translation termination and NMD on the human transcriptome and reveal novel determinants of ribosome biogenesis regulation.
Keywords: GSPT1; UPF1; eRF3; nonsense-mediated mRNA decay; ribosome protein genes; translation termination; uORF.
Figures
References
-
- Jackson RJ, Hellen CUT, Pestova TV.. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
