Transcriptional characterisation of human lung cells identifies novel mesenchymal lineage markers
- PMID: 31619469
- PMCID: PMC8055172
- DOI: 10.1183/13993003.00746-2019
Transcriptional characterisation of human lung cells identifies novel mesenchymal lineage markers
Abstract
Rationale: The lung mesenchyme gives rise to multiple distinct lineages of cells in the mature respiratory system, including smooth muscle cells of the airway and vasculature. However, a thorough understanding of the specification and mesenchymal cell diversity in the human lung is lacking.
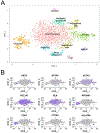
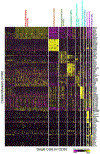
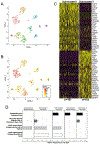
Methods: We completed single-cell RNA sequencing analysis of fetal human lung tissues. Canonical correlation analysis, clustering, cluster marker gene identification and t-distributed stochastic neighbour embedding representation was performed in Seurat. Cell populations were annotated using ToppFun. Immunohistochemistry and in situ hybridisation were used to validate spatiotemporal gene expression patterns for key marker genes.
Results: We identified molecularly distinct populations representing "committed" fetal human lung endothelial cells, pericytes and smooth muscle cells. Early endothelial lineages expressed "classic" endothelial cell markers (platelet endothelial cell adhesion molecule/CD31 and claudin 5), while pericytes expressed platelet-derived growth factor receptor-β, Thy-1 membrane glycoprotein and basement membrane molecules (collagen IV, laminin and proteoglycans). We observed a large population of "nonspecific" human lung mesenchymal progenitor cells characterised by expression of collagen I and multiple elastin fibre genes (ELN, MFAP2 and FBN1). We closely characterised the diversity of mesenchymal lineages defined by α2-smooth muscle actin (ACTA2) expression. Two cell populations, with the highest levels of ACTA2 transcriptional activity, expressed unique sets of markers associated with airway or vascular smooth muscle cells. Spatiotemporal analysis of these marker genes confirmed early and persistent spatial specification of airway (HHIP, MYLK and IGF1) and vascular (NTRK3 and MEF2C) smooth muscle cells in the developing human lung.
Conclusion: Our data suggest that specification of distinct airway and vascular smooth muscle cell phenotypes is established early in development and can be identified using the markers we provide.
Copyright ©ERS 2020.
Conflict of interest statement
Conflict of interest: S. Bhattacharya has nothing to disclose. Conflict of interest: T.J. Mariani has nothing to disclose. Conflict of interest: D. Al Alam has nothing to disclose. Conflict of interest: S. Danopoulos has nothing to disclose.
Figures





Comment in
-
Single cell analysis of human lung development: knowing what mesenchymal cells are and what they may be.Eur Respir J. 2020 Jan 23;55(1):1902327. doi: 10.1183/13993003.02327-2019. Print 2020 Jan. Eur Respir J. 2020. PMID: 31974123 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
