Regulation of lifespan by neural excitation and REST
- PMID: 31619788
- PMCID: PMC6893853
- DOI: 10.1038/s41586-019-1647-8
Regulation of lifespan by neural excitation and REST
Abstract
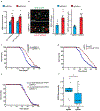
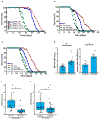
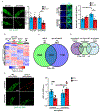
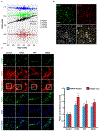
The mechanisms that extend lifespan in humans are poorly understood. Here we show that extended longevity in humans is associated with a distinct transcriptome signature in the cerebral cortex that is characterized by downregulation of genes related to neural excitation and synaptic function. In Caenorhabditis elegans, neural excitation increases with age and inhibition of excitation globally, or in glutamatergic or cholinergic neurons, increases longevity. Furthermore, longevity is dynamically regulated by the excitatory-inhibitory balance of neural circuits. The transcription factor REST is upregulated in humans with extended longevity and represses excitation-related genes. Notably, REST-deficient mice exhibit increased cortical activity and neuronal excitability during ageing. Similarly, loss-of-function mutations in the C. elegans REST orthologue genes spr-3 and spr-4 elevate neural excitation and reduce the lifespan of long-lived daf-2 mutants. In wild-type worms, overexpression of spr-4 suppresses excitation and extends lifespan. REST, SPR-3, SPR-4 and reduced excitation activate the longevity-associated transcription factors FOXO1 and DAF-16 in mammals and worms, respectively. These findings reveal a conserved mechanism of ageing that is mediated by neural circuit activity and regulated by REST.
Figures










Comment in
-
Moderation of neural excitation promotes longevity.Nature. 2019 Oct;574(7778):338-340. doi: 10.1038/d41586-019-02958-x. Nature. 2019. PMID: 31619782 No abstract available.
-
REST linked to longevity.Lab Anim (NY). 2020 Jan;49(1):22. doi: 10.1038/s41684-019-0448-x. Lab Anim (NY). 2020. PMID: 31819279 No abstract available.
References
-
- Satoh A, Imai S-i, Guarente L. The brain, sirtuins, and ageing. Nat Rev Neurosci. 2017;18(6):362–74. - PubMed
-
- Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402(6763):804–9. - PubMed
-
- Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41(1):45–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH113279/MH/NIMH NIH HHS/United States
- U01 AG046152/AG/NIA NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- R01 EY024376/EY/NEI NIH HHS/United States
- EY011930/NH/NIH HHS/United States
- K99 AG050830/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- R01 GM072551/GM/NIGMS NIH HHS/United States
- P50 MH084053/MH/NIMH NIH HHS/United States
- DP1 OD006849/OD/NIH HHS/United States
- P50 MH066392/MH/NIMH NIH HHS/United States
- R01 MH075916/MH/NIMH NIH HHS/United States
- EY024376/NH/NIH HHS/United States
- T32 AG000222/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- R01AG17917/NH/NIH HHS/United States
- R01 EY011930/EY/NEI NIH HHS/United States
- R01 MH097276/MH/NIMH NIH HHS/United States
- P01 AG002219/AG/NIA NIH HHS/United States
- R01 MH093725/MH/NIMH NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- DP1 AG044161/AG/NIA NIH HHS/United States
- R01AG36836/NH/NIH HHS/United States
- U01AG46152/NH/NIH HHS/United States
- R01 MH080405/MH/NIMH NIH HHS/United States
- R01 AG026651/AG/NIA NIH HHS/United States
- R01 AG036836/AG/NIA NIH HHS/United States
- R01 AG046174/AG/NIA NIH HHS/United States
- R01 MH085542/MH/NIMH NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

