Identification of Transgene-Free CRISPR-Edited Plants of Rice, Tomato, and Arabidopsis by Monitoring DsRED Fluorescence in Dry Seeds
- PMID: 31620160
- PMCID: PMC6759815
- DOI: 10.3389/fpls.2019.01150
Identification of Transgene-Free CRISPR-Edited Plants of Rice, Tomato, and Arabidopsis by Monitoring DsRED Fluorescence in Dry Seeds
Abstract
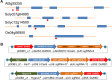
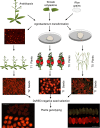
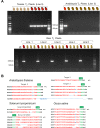
Efficient elimination of the editing machinery remains a challenge in plant biotechnology after genome editing to minimize the probability of off-target mutations, but it is also important to deliver end users with edited plants free of foreign DNA. Using the modular cloning system Golden Braid, we have included a fluorescence-dependent transgene monitoring module to the genome-editing tool box. We have tested this approach in Solanum lycopersicum, Oryza sativa, and Arabidopsis thaliana. We demonstrate that DsRED fluorescence visualization works efficiently in dry seeds as marker for the detection of the transgene in the three species allowing an efficient method for selecting transgene-free dry seeds. In the first generation of DsRED-free CRISPR/Cas9 null segregants, we detected gene editing of selected targets including homozygous mutants for the plant species tested. We demonstrate that this strategy allows rapid selection of transgene-free homozygous edited crop plants in a single generation after in vitro transformation.
Keywords: Arabidopsis thaliana; CRISPR/Cas9; DsRED; Oryza sativa; Solanum lycopersicum; genome editing.
Copyright © 2019 Aliaga-Franco, Zhang, Presa, Srivastava, Granell, Alabadí, Sadanandom, Blázquez and Minguet.
Figures
Similar articles
-
Highly efficient generation of bacterial leaf blight-resistant and transgene-free rice using a genome editing and multiplexed selection system.BMC Plant Biol. 2021 Apr 24;21(1):197. doi: 10.1186/s12870-021-02979-7. BMC Plant Biol. 2021. PMID: 33894749 Free PMC article.
-
H2O2-Based Method for Rapid Detection of Transgene-Free Rice Plants from Segregating CRISPR/Cas9 Genome-Edited Progenies.Int J Mol Sci. 2019 Aug 9;20(16):3885. doi: 10.3390/ijms20163885. Int J Mol Sci. 2019. PMID: 31404948 Free PMC article.
-
The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation.Plant Biotechnol J. 2014 Aug;12(6):797-807. doi: 10.1111/pbi.12200. Epub 2014 May 23. Plant Biotechnol J. 2014. PMID: 24854982
-
Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants.aBIOTECH. 2019 Dec 3;1(1):88-96. doi: 10.1007/s42994-019-00013-x. eCollection 2020 Jan. aBIOTECH. 2019. PMID: 36305007 Free PMC article. Review.
-
The CRISPR/Cas9 system and its applications in crop genome editing.Crit Rev Biotechnol. 2019 May;39(3):321-336. doi: 10.1080/07388551.2018.1554621. Epub 2019 Jan 15. Crit Rev Biotechnol. 2019. PMID: 30646772 Review.
Cited by
-
Current status and prospects of plant genome editing in Australia.In Vitro Cell Dev Biol Plant. 2021;57(4):574-583. doi: 10.1007/s11627-021-10188-y. Epub 2021 May 24. In Vitro Cell Dev Biol Plant. 2021. PMID: 34054265 Free PMC article. Review.
-
A bifunctional selectable marker for wheat transformation contributes to the characterization of male-sterile phenotype induced by a synthetic Ms2 gene.Plant Cell Rep. 2023 May;42(5):895-907. doi: 10.1007/s00299-023-02998-8. Epub 2023 Mar 3. Plant Cell Rep. 2023. PMID: 36867203
-
A Prospective Review on Selectable Marker-Free Genome Engineered Rice: Past, Present and Future Scientific Realm.Front Genet. 2022 Jun 9;13:882836. doi: 10.3389/fgene.2022.882836. eCollection 2022. Front Genet. 2022. PMID: 35754795 Free PMC article. Review.
-
Engineering drought tolerance in plants through CRISPR/Cas genome editing.3 Biotech. 2020 Sep;10(9):400. doi: 10.1007/s13205-020-02390-3. Epub 2020 Aug 19. 3 Biotech. 2020. PMID: 32864285 Free PMC article. Review.
-
Ares-GT: Design of guide RNAs targeting multiple genes for CRISPR-Cas experiments.PLoS One. 2020 Oct 21;15(10):e0241001. doi: 10.1371/journal.pone.0241001. eCollection 2020. PLoS One. 2020. PMID: 33085710 Free PMC article.
References
-
- Abbas M., Hernandez-Garcia J., Blanco-Tourinan N., Aliaga N., Minguet E. G., Alabadi D., et al. (2018. a). Reduction of indole-3-acetic acid methyltransferase activity compensates for high-temperature male sterility in Arabidopsis. Plant Biotechnol. J. 16, 272–279. 10.1111/pbi.12768 - DOI - PMC - PubMed

