Immune Landscape of Invasive Ductal Carcinoma Tumor Microenvironment Identifies a Prognostic and Immunotherapeutically Relevant Gene Signature
- PMID: 31620363
- PMCID: PMC6759595
- DOI: 10.3389/fonc.2019.00903
Immune Landscape of Invasive Ductal Carcinoma Tumor Microenvironment Identifies a Prognostic and Immunotherapeutically Relevant Gene Signature
Abstract
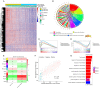
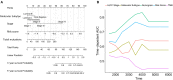
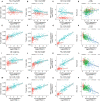
Background: Invasive ductal carcinoma (IDC) is a clinically and molecularly distinct disease. Tumor microenvironment (TME) immune phenotypes play crucial roles in predicting clinical outcomes and therapeutic efficacy. Method: In this study, we depict the immune landscape of IDC by using transcriptome profiling and clinical characteristics retrieved from The Cancer Genome Atlas (TCGA) data portal. Immune cell infiltration was evaluated via single-sample gene set enrichment (ssGSEA) analysis and systematically correlated with genomic characteristics and clinicopathological features of IDC patients. Furthermore, an immune signature was constructed using the least absolute shrinkage and selection operator (LASSO) Cox regression algorithm. A random forest algorithm was applied to identify the most important somatic gene mutations associated with the constructed immune signature. A nomogram that integrated clinicopathological features with the immune signature to predict survival probability was constructed by multivariate Cox regression. Results: The IDC were clustered into low immune infiltration, intermediate immune infiltration, and high immune infiltration by the immune landscape. The high infiltration group had a favorable survival probability compared with that of the low infiltration group. The low-risk score subtype identified by the immune signature was characterized by T cell-mediated immune activation. Additionally, activation of the interferon-α response, interferon-γ response, and TNF-α signaling via the NFκB pathway was observed in the low-risk score subtype, which indicated T cell activation and may be responsible for significantly favorable outcomes in IDC patients. A random forest algorithm identified the most important somatic gene mutations associated with the constructed immune signature. Furthermore, a nomogram that integrated clinicopathological features with the immune signature to predict survival probability was constructed, revealing that the immune signature was an independent prognostic biomarker. Finally, the relationship of VEGFA, PD1, PDL-1, and CTLA-4 expression with the immune infiltration landscape and the immune signature was analyzed to interpret the responses of IDC patients to immunotherapy. Conclusion: Taken together, we performed a comprehensive evaluation of the immune landscape of IDC and constructed an immune signature related to the immune landscape. This analysis of TME immune infiltration landscape has shed light on how IDC respond to immunotherapy and may guide the development of novel drug combination strategies.
Keywords: immune checkpoint inhibitor; immune landscape; immune signature; invasive ductal carcinoma; survival.
Copyright © 2019 Bao, Shi, Zhang, Xin, Li, Zhao and Wang.
Figures





References
-
- Eble JN, Tavassoli FA, Devilee P. Pathology and Genetics of Tumours of the Breast and Female Genital Organs. Iarc; (2003).
-
- Smid M, Rodríguez-González FG, Sieuwerts AM, Salgado R, Prager-Van der Smissen WJ, van Der Vlugt-Daane M, et al. . Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat Commun. (2016) 7:12910. 10.1038/ncomms12910 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

