OleD Loki as a Catalyst for Hydroxamate Glycosylation
- PMID: 31621997
- PMCID: PMC7124993
- DOI: 10.1002/cbic.201900601
OleD Loki as a Catalyst for Hydroxamate Glycosylation
Abstract
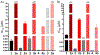
Herein we describe the ability of the permissive glycosyltransferase (GT) OleD Loki to convert a diverse set of >15 histone deacetylase (HDAC) inhibitors (HDACis) into their corresponding hydroxamate glycosyl esters. Representative glycosyl esters were subsequently evaluated in assays for cancer cell line cytotoxicity, chemical and enzymatic stability, and axolotl embryo tail regeneration. Computational substrate docking models were predictive of enzyme-catalyzed turnover and suggest certain HDACis may form unproductive, potentially inhibitory, complexes with GTs.
Keywords: HDAC; glucosylation; glycosyltransferase; histone deacetylase.
© 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of Interest
J.S.T. is a co-founder of Centrose (Madison, WI, USA).
Figures



Similar articles
-
OleD Loki as a Catalyst for Tertiary Amine and Hydroxamate Glycosylation.Chembiochem. 2017 Feb 16;18(4):363-367. doi: 10.1002/cbic.201600676. Epub 2017 Jan 9. Chembiochem. 2017. PMID: 28067448 Free PMC article.
-
Design, Synthesis and Biological Evaluation of Novel Coumarin-Based Hydroxamate Derivatives as Histone Deacetylase (Hdac) Inhibitors with Antitumor Activities.Molecules. 2019 Jul 15;24(14):2569. doi: 10.3390/molecules24142569. Molecules. 2019. PMID: 31311163 Free PMC article.
-
Improved antiproliferative activity of 1,3,4-thiadiazole-containing histone deacetylase (HDAC) inhibitors by introduction of the heteroaromatic surface recognition motif.Bioorg Med Chem. 2014 Nov 1;22(21):5766-75. doi: 10.1016/j.bmc.2014.09.039. Epub 2014 Sep 28. Bioorg Med Chem. 2014. PMID: 25311567
-
The Development Process: from SAHA to Hydroxamate HDAC Inhibitors with Branched CAP Region and Linear Linker.Chem Biodivers. 2020 Jan;17(1):e1900427. doi: 10.1002/cbdv.201900427. Epub 2019 Dec 23. Chem Biodivers. 2020. PMID: 31793143 Review.
-
Structure-activity relationships of hydroxamate-based histone deacetylase-8 inhibitors: reality behind anticancer drug discovery.Future Med Chem. 2017 Dec;9(18):2211-2237. doi: 10.4155/fmc-2017-0130. Epub 2017 Nov 28. Future Med Chem. 2017. PMID: 29182018 Review.
Cited by
-
Design of a chimeric glycosyltransferase OleD for the site-specific O-monoglycosylation of 3-hydroxypyridine in nosiheptide.Microb Biotechnol. 2023 Oct;16(10):1971-1984. doi: 10.1111/1751-7915.14332. Epub 2023 Aug 22. Microb Biotechnol. 2023. PMID: 37606280 Free PMC article.
-
Sugar-Pirating as an Enabling Platform for the Synthesis of 4,6-Dideoxyhexoses.J Am Chem Soc. 2020 May 20;142(20):9389-9395. doi: 10.1021/jacs.9b13766. Epub 2020 May 7. J Am Chem Soc. 2020. PMID: 32330028 Free PMC article.
References
-
- Elshahawi SI, Shaaban KA, Kharel MK, Thorson JS, Chem. Soc. Rev 2015, 44, 7591–7697; - PMC - PubMed
- Gloster TM, Curr. Opin. Struct. Biol 2014, 28, 131–141; - PMC - PubMed
- Breton C, Fournel-Gigleux S, Palcic MM, Curr. Opin. Struct. Biol 2012, 22, 540–549; - PubMed
- Singh S, Phillips GN Jr., Thorson JS, Nat. Prod. Rep 2012, 29, 1201–1237; - PMC - PubMed
- Chang A, Singh S, Phillips GN Jr., Thorson JS, Curr. Opin. Biotechnol 2011, 22, 800–808; - PMC - PubMed
- Gantt RW, Peltier-Pain P, Thorson JS, Nat. Prod. Rep 2011, 28, 1811–1853; - PubMed
- Palcic MM, Curr. Opin. Chem. Biol 2011, 15, 226–233; - PubMed
- Roychoudhury R, Pohl NL, Curr. Opin. Chem. Biol 2010, 14, 168–173; - PubMed
- Lairson L, Henrissat B, Davies G, Withers S, Annu. Rev. Biochem 2008, 77, 521–555; - PubMed
- Bowles D, Lim E, Poppenberger B, Vaistij F, Annu. Rev. Plant Biol 2006, 57, 567–597. - PubMed
-
- Wu CZ, Jang JH, Woo M, Ahn JS, Kim JS, Hong Y, Appl. Environ. Microbiol 2012, 78, 7680–7686; - PMC - PubMed
- Ahn B, Kim B, Jeon Y, Lee E, Lim Y, Ahn J, J. Microbiol. Biotechnol 2009, 19, 387–390; - PubMed
- Méndez C, Salas J, Ernst Schering Res. Found. Workshop 2005, 51, 127–146; - PubMed
- Lu W, Oberthur M, Leimkuhler C, Tao J, Kahne D, Walsh C, Proc. Natl. Acad. Sci. USA 2004, 101, 4390–4395; - PMC - PubMed
- Rodriguez L, Aguirrezabalaga I, Allende N, Brana A, Mendez C, Salas J, Chem. Biol 2002, 9, 721–729. - PubMed
-
- Guo Z, Li J, Qin H, Wang M, Lv X, Li X, Chen Y, Angew. Chem. Int. Ed 2015, 54, 5175–5178; - PubMed
- Angew. Chem 2015, 127, 5264–5267;
- Gawthorne JA, Tan NY, Bailey UM, Davis MR, Wong LW, Naidu R, Fox KL, Jennings MP, Schulz BL, Biochem. Biophys. Res. Commun 2014, 445, 633–638; - PubMed
- Naegeli A, Michaud G, Schubert M, Lin CW, Lizak C, Darbre T, Reymond JL, Aebi M, J. Biol. Chem 2014, 289, 24521–24532; - PMC - PubMed
- Magarvey NA, Haltli B, He M, Greenstein M, Hucul JA, Antimicrob. Agents Chemother 2006, 50, 2167–2177; - PMC - PubMed
- Gao Q, Zhang C, Blanchard S, Thorson JS, Chem. Biol 2006, 13, 733–743; - PubMed
- Sánchez C, Méndez C, Salas JA, Nat. Prod. Rep 2006, 23, 1007–1045; - PubMed
- Zhang C, Albermann C, Fu X, Peters NR, Chisholm JD, Zhang G, Gilbert EJ, Wang PG, Van Vranken DL, Thorson JS, ChemBioChem 2006, 7, 795–804; - PubMed
- Salas AP, Zhu L, Sáanchez C, Braña AF, Rohr J, Méendez C, Salas JA, Mol. Microbiol 2005, 58, 17–27; - PMC - PubMed
- Hyun CG, Bililign T, Liao J, Thorson JS, ChemBioChem 2003, 4, 114–117; - PubMed
- Sáanchez C, Butovich IA, Braña AF, Rohr J, Méendez C, Salas JA, Chem. Biol 2002, 9, 519–531. - PubMed
-
- Calce E, Digilio G, Menchise V, Saviano M, De Luca S, Chem. Eur. J 2018, 24, 6231–6238; - PubMed
- Kopycki J, Wieduwild E, Kohlschmidt J, Brandt W, Stepanova AN, Alonso JM, Pedras MS, Abel S, Grubb CD, Biochem. J 2013, 450, 37–46; - PubMed
- Jahn M, Marles J, Warren R, Withers S, Angew. Chem. Int. Ed 2003, 42, 352–354; - PubMed
- Angew. Chem 2003, 115, 366–368.
-
- Chen D, Chen R, Xie K, Yue T, Zhang X, Ye F, Dai J, Org. Lett 2018, 20, 1634–1637; - PubMed
- Chen D, Sun L, Chen R, Xie K, Yang L, Dai J, Chem. Eur. J 2016, 22, 5873–5877; - PubMed
- Chen D, Chen R, Wang R, Li J, Xie K, Bian C, Sun L, Zhang X, Liu J, Yang L, Ye F, Yu X, Dai J, Angew. Chem. Int. Ed 2015, 54, 12678–12682; - PubMed
- Angew. Chem 2015, 127, 12869–12873;
- Foshag D, Campbell C, Pawelek PD, Biochim. Biophys. Acta 2014, 1844, 1619–1630; - PubMed
- Gutmann A, Krump C, Bungaruang L, Nidetzky B, Chem. Commun 2014, 50, 5465–5468; - PubMed
- Li L, Wang P, Tang Y, J. Antibiot 2014, 67, 65–70; - PubMed
- Wang F, Zhou M, Singh S, Yennamalli RM, Bingman CA, Thorson JS, Phillips GN Jr., Proteins 2013, 81, 1277–1282; - PMC - PubMed
- Gutmann A, Nidetzky B, Angew. Chem. Int. Ed 2012, 51, 12879–12883; - PubMed
- Angew. Chem 2012, 124, 13051–13056;
- Härle J, Günther S, Lauinger B, Weber M, Kammerer B, Zechel DL, Luzhetskyy A, Bechthold A, Chem. Biol 2011, 18, 520–530; - PubMed
- Mittler M, Bechthold A, Schulz GE, J. Mol. Biol 2007, 372, 67–76; - PubMed
- Baig I, Kharel M, Kobylyanskyy A, Zhu L, Rebets Y, Ostash B, Luzhetskyy A, Bechthold A, Fedorenko VA, Rohr J, Angew. Chem. Int. Ed 2006, 45, 7842–7846; - PMC - PubMed
- Angew. Chem 2006, 118, 8006–8010;
- Liu T, Kharel MK, Fischer C, McCormick A, Rohr J, ChemBioChem 2006, 7, 1070–1077; - PMC - PubMed
- Fischbach MA, Lin H, Liu DR, Walsh CT, Proc. Natl. Acad. Sci. USA 2005, 102, 571–576; - PMC - PubMed
- Bililign T, Hyun CG, Williams JS, Czisny AM, Thorson JS, Chem. Biol 2004, 11, 959–969. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

