Multivalent Ultrasensitive Interfacing of Supramolecular 1D Nanoplatforms
- PMID: 31622094
- PMCID: PMC6856958
- DOI: 10.1021/jacs.9b05629
Multivalent Ultrasensitive Interfacing of Supramolecular 1D Nanoplatforms
Abstract
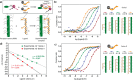
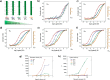
Multivalent display on linear platforms is used by many biomolecular systems to effectively interact with their corresponding binding partners in a dose-responsive and ultrasensitive manner appropriate to the biological system at hand. Synthetic supramolecular multivalent displays offer a matching approach for the modular and bottom-up construction and systematic study of dynamic 1D materials. Fundamental studies into multivalent interactions between such linear, 1D materials have been lacking because of the absence of appropriate modular nanoplatforms. In this work we interfaced two synthetic multivalent linear nanoplatforms based on a dynamic supramolecular polymer, formed by hybrid discotic-oligonucleotide monomers, and a series of complementary DNA-duplex-based multivalent ligands, also with appended short oligonucleotides. The combination of these two multivalent nanoplatforms provides for the first time entry to study multivalent effects in dynamic 1D systems, of relevance for the conceptual understanding of multivalency in biology and for the generation of novel multivalent biomaterials. Together the two nanoscaffolds provide easy access to libraries of multivalent ligands with tunable affinities. The DNA scaffold allows for exact control over valency and spatial ligand distribution, and the discotic supramolecular polymer allows for dynamic adaptation and control over receptor density. The interaction between the two nanoplatforms was studied as a function of ligand interaction strength, valency, and density. Usage of the enhancement parameter β allowed quantification of the effects of ligand valency and affinity. The results reveal a generalized principle of additive binding increments. Receptor density is shown to be crucially and nonlinearly correlated to complex formation, leading to ultrasensitive responses. The results reveal that, not unlike biomolecular signaling, high density multivalent display of receptors is crucial for functionally increased affinities.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Li P.; Banjade S.; Cheng H.-C.; Kim S.; Chen B.; Guo L.; Llaguno M.; Hollingsworth J. V.; King D. S.; Banani S. F.; Russo P. S.; Jiang Q.-X.; Nixon B. T.; Rosen M. K. Phase Transitions in the Assembly of Multivalent Signalling Proteins. Nature 2012, 483, 336–340. 10.1038/nature10879. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

