Ubiquitin Ligases Involved in the Regulation of Wnt, TGF-β, and Notch Signaling Pathways and Their Roles in Mouse Development and Homeostasis
- PMID: 31623112
- PMCID: PMC6826584
- DOI: 10.3390/genes10100815
Ubiquitin Ligases Involved in the Regulation of Wnt, TGF-β, and Notch Signaling Pathways and Their Roles in Mouse Development and Homeostasis
Abstract
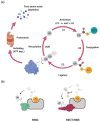
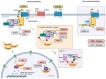
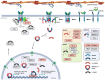
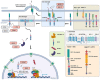
The Wnt, TGF-β, and Notch signaling pathways are essential for the regulation of cellular polarity, differentiation, proliferation, and migration. Differential activation and mutual crosstalk of these pathways during animal development are crucial instructive forces in the initiation of the body axis and the development of organs and tissues. Due to the ability to initiate cell proliferation, these pathways are vulnerable to somatic mutations selectively producing cells, which ultimately slip through cellular and organismal checkpoints and develop into cancer. The architecture of the Wnt, TGF-β, and Notch signaling pathways is simple. The transmembrane receptor, activated by the extracellular stimulus, induces nuclear translocation of the transcription factor, which subsequently changes the expression of target genes. Nevertheless, these pathways are regulated by a myriad of factors involved in various feedback mechanisms or crosstalk. The most prominent group of regulators is the ubiquitin-proteasome system (UPS). To open the door to UPS-based therapeutic manipulations, a thorough understanding of these regulations at a molecular level and rigorous confirmation in vivo are required. In this quest, mouse models are exceptional and, thanks to the progress in genetic engineering, also an accessible tool. Here, we reviewed the current understanding of how the UPS regulates the Wnt, TGF-β, and Notch pathways and we summarized the knowledge gained from related mouse models.
Keywords: cancer; gene inactivation; mouse model; ubiquitin–proteasome system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

