Pathogenetic Impact of Bacterial-Fungal Interactions
- PMID: 31623187
- PMCID: PMC6843596
- DOI: 10.3390/microorganisms7100459
Pathogenetic Impact of Bacterial-Fungal Interactions
Abstract
Polymicrobial infections are of paramount importance because of the potential severity of clinical manifestations, often associated with increased resistance to antimicrobial treatment. The intricate interplay with the host and the immune system, and the impact on microbiome imbalance, are of importance in this context. The equilibrium of microbiota in the human host is critical for preventing potential dysbiosis and the ensuing development of disease. Bacteria and fungi can communicate via signaling molecules, and produce metabolites and toxins capable of modulating the immune response or altering the efficacy of treatment. Most of the bacterial-fungal interactions described to date focus on the human fungal pathogen Candida albicans and different bacteria. In this review, we discuss more than twenty different bacterial-fungal interactions involving several clinically important human pathogens. The interactions, which can be synergistic or antagonistic, both in vitro and in vivo, are addressed with a focus on the quorum-sensing molecules produced, the response of the immune system, and the impact on clinical outcome.
Keywords: bacterial–fungal interactions; immune response; in vivo models; microbiome; molecules.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
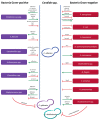
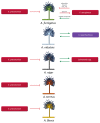
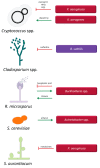
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

