Global Burden of Colistin-Resistant Bacteria: Mobilized Colistin Resistance Genes Study (1980-2018)
- PMID: 31623244
- PMCID: PMC6843232
- DOI: 10.3390/microorganisms7100461
Global Burden of Colistin-Resistant Bacteria: Mobilized Colistin Resistance Genes Study (1980-2018)
Abstract
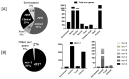
Colistin is considered to be an antimicrobial of last-resort for the treatment of multidrug-resistant Gram-negative bacterial infections. The recent global dissemination of mobilized colistin resistance (mcr) genes is an urgent public health threat. An accurate estimate of the global prevalence of mcr genes, their reservoirs and the potential pathways for human transmission are required to implement control and prevention strategies, yet such data are lacking. Publications from four English (PubMed, Scopus, the Cochrane Database of Systematic Reviews and Web of Science) and two Chinese (CNKI and WANFANG) databases published between 18 November 2015 and 30 December 2018 were identified. In this systematic review and meta-analysis, the prevalence of mcr genes in bacteria isolated from humans, animals, the environment and food products were investigated. A total of 974 publications were identified. 202 observational studies were included in the systematic review and 71 in the meta-analysis. mcr genes were reported from 47 countries across six continents and the overall average prevalence was 4.7% (0.1-9.3%). China reported the highest number of mcr-positive strains. Pathogenic Escherichia coli (54%), isolated from animals (52%) and harboring an IncI2 plasmid (34%) were the bacteria with highest prevalence of mcr genes. The estimated prevalence of mcr-1 pathogenic E. coli was higher in food-animals than in humans and food products, which suggests a role for foodborne transmission. This study provides a comprehensive assessment of prevalence of the mcr gene by source, organism, genotype and type of plasmid.
Keywords: Escherichia coli; antibiotic resistance; food-chain; meta-analysis; mobilized colistin resistance (mcr) genes; reservoir.
Conflict of interest statement
All the authors declare no competing interests.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources

