Analysis of miRNAs in the Heads of Different Castes of the Bumblebee Bombus lantschouensis (Hymenoptera: Apidae)
- PMID: 31623265
- PMCID: PMC6835379
- DOI: 10.3390/insects10100349
Analysis of miRNAs in the Heads of Different Castes of the Bumblebee Bombus lantschouensis (Hymenoptera: Apidae)
Abstract
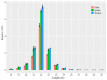
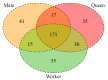
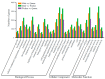
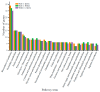
Bumblebees are important insect pollinators for many wildflowers and crops. MicroRNAs (miRNAs) are endogenous non-coding small RNAs that regulate different biological functions in insects. In this study, the miRNAs in the heads of the three castes of the bumblebee Bombus lantschouensis were identified and characterized by small RNA deep sequencing. The significant differences in the expression of miRNAs and their target genes were analyzed. The results showed that the length of the small RNA reads from males, queens, and workers was distributed between 18 and 30 nt, with a peak at 22 nt. A total of 364 known and 89 novel miRNAs were identified from the heads of the three castes. The eight miRNAs with the highest expressed levels in males, queens, and workers were identical, although the order of these miRNAs based on expression differed. The male vs. queen, male vs. worker, and worker vs. queen comparisons identified nine, fourteen, and four miRNAs with significant differences in expression, respectively. The different castes were clustered based on the differentially expressed miRNAs (DE miRNAs), and the expression levels of the DE miRNAs obtained by RT-qPCR were consistent with the read counts obtained through Solexa sequencing. The putative target genes of these DE miRNAs were enriched in 29 Gene Ontology (GO) terms, and catalytic activity was the most enriched GO term, as demonstrated by its association with 2837 target genes in the male vs. queen comparison, 3535 target genes in the male vs. worker comparison, and 2185 target genes in the worker vs. queen comparison. This study highlights the characteristics of the miRNAs in the three B. lantschouensis castes and will aid further studies on the functions of miRNAs in bumblebees.
Keywords: Bombus lantschouensis; Solexa sequencing; microRNA; pollinators.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Characterization of a Vitellogenin Receptor in the Bumblebee, Bombus lantschouensis (Hymenoptera, Apidae).Insects. 2019 Dec 12;10(12):445. doi: 10.3390/insects10120445. Insects. 2019. PMID: 31842304 Free PMC article.
-
Comparison of Morphological Characteristics of Antennae and Antennal Sensilla among Four Species of Bumblebees (Hymenoptera: Apidae).Insects. 2023 Feb 26;14(3):232. doi: 10.3390/insects14030232. Insects. 2023. PMID: 36975917 Free PMC article.
-
Host choice in the phoretic mite Parasitellus fucorum (Mesostigmata: Parasitidae): which bumblebee caste is the best?Oecologia. 1998 Jul;115(3):385-390. doi: 10.1007/s004420050532. Oecologia. 1998. PMID: 28308431
-
Transcript expression bias of phosphatidylethanolamine binding protein gene in bumblebee, Bombus lantschouensis (Hymenoptera: Apidae).Gene. 2017 Sep 5;627:290-297. doi: 10.1016/j.gene.2017.06.041. Epub 2017 Jun 21. Gene. 2017. PMID: 28647560
-
Workers dominate male production in the neotropical bumblebee Bombus wilmattae (Hymenoptera: Apidae).Front Zool. 2011 Jun 8;8(1):13. doi: 10.1186/1742-9994-8-13. Front Zool. 2011. PMID: 21651814 Free PMC article.
Cited by
-
Age and Behavior-Dependent Differential miRNAs Expression in the Hypopharyngeal Glands of Honeybees (Apis mellifera L.).Insects. 2021 Aug 26;12(9):764. doi: 10.3390/insects12090764. Insects. 2021. PMID: 34564204 Free PMC article.
-
Effects of Dinotefuran on Brain miRNA Expression Profiles in Young Adult Honey Bees (Hymenopptera: Apidae).J Insect Sci. 2021 Jan 1;21(1):3. doi: 10.1093/jisesa/ieaa131. J Insect Sci. 2021. PMID: 33400795 Free PMC article.
References
-
- Huang J., An J. Species diversity, pollination application and strategy for conservation of the bumblebees of China. Biodivers. Sci. 2018;26:486–497. doi: 10.17520/biods.2018068. - DOI
-
- Velthuis H.H.W., van Doorn A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie. 2006;37:421–451. doi: 10.1051/apido:2006019. - DOI
-
- Thorp R.W., Horning D.S., Dunning L.L. Bumble bees and cuckoo bumble bees of California (Hyenoptera, Apidae) Bull. Calif. Insect Surv. 1983;23:1–79.
LinkOut - more resources
Full Text Sources
Research Materials

