The static and dynamic structural heterogeneities of B-DNA: extending Calladine-Dickerson rules
- PMID: 31624840
- PMCID: PMC6868377
- DOI: 10.1093/nar/gkz905
The static and dynamic structural heterogeneities of B-DNA: extending Calladine-Dickerson rules
Abstract
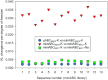
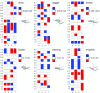
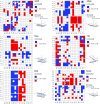
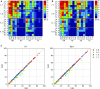
We present a multi-laboratory effort to describe the structural and dynamical properties of duplex B-DNA under physiological conditions. By processing a large amount of atomistic molecular dynamics simulations, we determine the sequence-dependent structural properties of DNA as expressed in the equilibrium distribution of its stochastic dynamics. Our analysis includes a study of first and second moments of the equilibrium distribution, which can be accurately captured by a harmonic model, but with nonlocal sequence-dependence. We characterize the sequence-dependent choreography of backbone and base movements modulating the non-Gaussian or anharmonic effects manifested in the higher moments of the dynamics of the duplex when sampling the equilibrium distribution. Contrary to prior assumptions, such anharmonic deformations are not rare in DNA and can play a significant role in determining DNA conformation within complexes. Polymorphisms in helical geometries are particularly prevalent for certain tetranucleotide sequence contexts and are always coupled to a complex network of coordinated changes in the backbone. The analysis of our simulations, which contain instances of all tetranucleotide sequences, allow us to extend Calladine-Dickerson rules used for decades to interpret the average geometry of DNA, leading to a set of rules with quantitative predictive power that encompass nonlocal sequence-dependence and anharmonic fluctuations.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Neidle S. Principles of Nucleic Acid Structure. 2008; London: Academic Press.
-
- Balaceanu A., Pasi M., Dans P.D., Hospital A., Lavery R., Orozco M.. The role of unconventional hydrogen bonds in determining BII propensities in B-DNA. J. Phys. Chem. Lett. 2017; 8:21–28. - PubMed

