Differentially expressed autophagy-related genes are potential prognostic and diagnostic biomarkers in clear-cell renal cell carcinoma
- PMID: 31626592
- PMCID: PMC6834403
- DOI: 10.18632/aging.102368
Differentially expressed autophagy-related genes are potential prognostic and diagnostic biomarkers in clear-cell renal cell carcinoma
Abstract
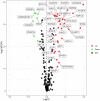
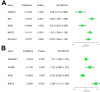
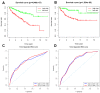
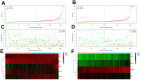
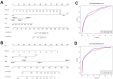
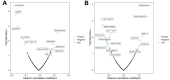
We examined the role of differentially expressed autophagy-related genes (DEARGs) in clear cell Renal Cell Carcinoma (ccRCC) using high-throughput RNA-seq data from The Cancer Genome Atlas (TCGA). Cox regression analyses showed that 5 DEARGs (PRKCQ, BID, BAG1, BIRC5, and ATG16L2) correlated with overall survival (OS) and 4 DEARGs (EIF4EBP1, BAG1, ATG9B, and BIRC5) correlated with disease-free survival (DFS) in ccRCC patients. Multivariate Cox regression analysis using the OS and DFS prognostic risk models showed that expression of the nine DEARGs accurately and independently predicted the risk of disease recurrence or progression in ccRCC patients (area under curve or AUC values > 0.70; all p < 0.05). Moreover, the DEARGs accurately distinguished healthy individuals from ccRCC patients based on receiver operated characteristic (ROC) analyses (area under curve or AUC values > 0.60), suggesting their potential as diagnostic biomarkers for ccRCC. The expression of DEARGs also correlated with the drug sensitivity of ccRCC cell lines. The ccRCC cell lines were significantly sensitive to Sepantronium bromide, a drug that targets BIRC5. This makes BIRC5 a potential therapeutic target for ccRCC. Our study thus demonstrates that DEARGs are potential diagnostic and prognostic biomarkers and therapeutic targets in ccRCC.
Keywords: TCGA database; autophagy; autophagy-related genes; ccRCC; prognosis.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

