Generation of Blastocyst-like Structures from Mouse Embryonic and Adult Cell Cultures
- PMID: 31626770
- PMCID: PMC7359735
- DOI: 10.1016/j.cell.2019.09.029
Generation of Blastocyst-like Structures from Mouse Embryonic and Adult Cell Cultures
Abstract
A single mouse blastomere from an embryo until the 8-cell stage can generate an entire blastocyst. Whether laboratory-cultured cells retain a similar generative capacity remains unknown. Starting from a single stem cell type, extended pluripotent stem (EPS) cells, we established a 3D differentiation system that enabled the generation of blastocyst-like structures (EPS-blastoids) through lineage segregation and self-organization. EPS-blastoids resembled blastocysts in morphology and cell-lineage allocation and recapitulated key morphogenetic events during preimplantation and early postimplantation development in vitro. Upon transfer, some EPS-blastoids underwent implantation, induced decidualization, and generated live, albeit disorganized, tissues in utero. Single-cell and bulk RNA-sequencing analysis revealed that EPS-blastoids contained all three blastocyst cell lineages and shared transcriptional similarity with natural blastocysts. We also provide proof of concept that EPS-blastoids can be generated from adult cells via cellular reprogramming. EPS-blastoids provide a unique platform for studying early embryogenesis and pave the way to creating viable synthetic embryos by using cultured cells.
Keywords: EPS cells; EPS-blastoid; blastocyst; blastoids; extended pluripotent stem cells; implantation; reprogramming; totipotent.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
None
Figures

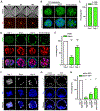

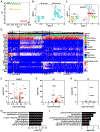
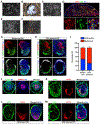
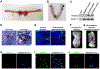
Comment in
-
A mouse blastocyst-like structure from scratch.Nat Methods. 2019 Dec;16(12):1207. doi: 10.1038/s41592-019-0674-6. Nat Methods. 2019. PMID: 31780830 No abstract available.
References
-
- Barton SC, Ferguson-Smith AC, Fundele R, and Surani MA (1991). Influence of paternally imprinted genes on development. Development 113, 679–687. - PubMed
-
- Beccari L, Moris N, Girgin M, Turner DA, Baillie-Johnson P, Cossy A-C, Lutolf MP, Duboule D, and Arias AM (2018). Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 2013 500:7462 562, 272–276. - PubMed
-
- Bedzhov I, Leung CY, Bialecka M, and Zernicka-Goetz M (2014a). In vitro culture of mouse blastocysts beyond the implantation stages. Nat Protoc 9, 2732–2739. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

