B-1a cells acquire their unique characteristics by bypassing the pre-BCR selection stage
- PMID: 31628339
- PMCID: PMC6802180
- DOI: 10.1038/s41467-019-12824-z
B-1a cells acquire their unique characteristics by bypassing the pre-BCR selection stage
Abstract
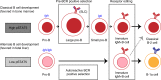
B-1a cells are long-lived, self-renewing innate-like B cells that predominantly inhabit the peritoneal and pleural cavities. In contrast to conventional B-2 cells, B-1a cells have a receptor repertoire that is biased towards bacterial and self-antigens, promoting a rapid response to infection and clearing of apoptotic cells. Although B-1a cells are known to primarily originate from fetal tissues, the mechanisms by which they arise has been a topic of debate for many years. Here we show that in the fetal liver versus bone marrow environment, reduced IL-7R/STAT5 levels promote immunoglobulin kappa gene recombination at the early pro-B cell stage. As a result, differentiating B cells can directly generate a mature B cell receptor (BCR) and bypass the requirement for a pre-BCR and pairing with surrogate light chain. This 'alternate pathway' of development enables the production of B cells with self-reactive, skewed specificity receptors that are peculiar to the B-1a compartment. Together our findings connect seemingly opposing lineage and selection models of B-1a cell development and explain how these cells acquire their unique properties.
Conflict of interest statement
The authors declare no competing interests.
Figures
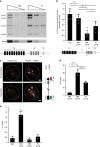


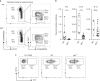


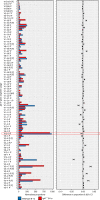

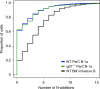
References
-
- Royston I, Majda JA, Baird SM, Meserve BL, Griffiths JC. Human T cell antigens defined by monoclonal antibodies: the 65,000-dalton antigen of T cells (T65) is also found on chronic lymphocytic leukemia cells bearing surface immunoglobulin. J. Immunol. 1980;125:725–731. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

