Altered DNA ligase activity in human disease
- PMID: 31630206
- PMCID: PMC7317150
- DOI: 10.1093/mutage/gez026
Altered DNA ligase activity in human disease
Abstract
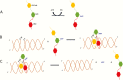
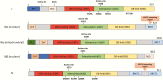
The joining of interruptions in the phosphodiester backbone of DNA is critical to maintain genome stability. These breaks, which are generated as part of normal DNA transactions, such as DNA replication, V(D)J recombination and meiotic recombination as well as directly by DNA damage or due to DNA damage removal, are ultimately sealed by one of three human DNA ligases. DNA ligases I, III and IV each function in the nucleus whereas DNA ligase III is the sole enzyme in mitochondria. While the identification of specific protein partners and the phenotypes caused either by genetic or chemical inactivation have provided insights into the cellular functions of the DNA ligases and evidence for significant functional overlap in nuclear DNA replication and repair, different results have been obtained with mouse and human cells, indicating species-specific differences in the relative contributions of the DNA ligases. Inherited mutations in the human LIG1 and LIG4 genes that result in the generation of polypeptides with partial activity have been identified as the causative factors in rare DNA ligase deficiency syndromes that share a common clinical symptom, immunodeficiency. In the case of DNA ligase IV, the immunodeficiency is due to a defect in V(D)J recombination whereas the cause of the immunodeficiency due to DNA ligase I deficiency is not known. Overexpression of each of the DNA ligases has been observed in cancers. For DNA ligase I, this reflects increased proliferation. Elevated levels of DNA ligase III indicate an increased dependence on an alternative non-homologous end-joining pathway for the repair of DNA double-strand breaks whereas elevated level of DNA ligase IV confer radioresistance due to increased repair of DNA double-strand breaks by the major non-homologous end-joining pathway. Efforts to determine the potential of DNA ligase inhibitors as cancer therapeutics are on-going in preclinical cancer models.
© The Author(s) 2019. Published by Oxford University Press on behalf of the UK Environmental Mutagen Society. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Ho C. K., Wang L. K., Lima C. D. and Shuman S (2004) Structure and mechanism of RNA ligase. Structure, 12, 327–339. - PubMed
-
- Shuman S. and Schwer B (1995) RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol., 17, 405–410. - PubMed
-
- Ahel I., Rass U., El-Khamisy S. F., Katyal S., Clements P. M., McKinnon P. J., Caldecott K. W. and West S. C (2006) The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature, 443, 713–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

